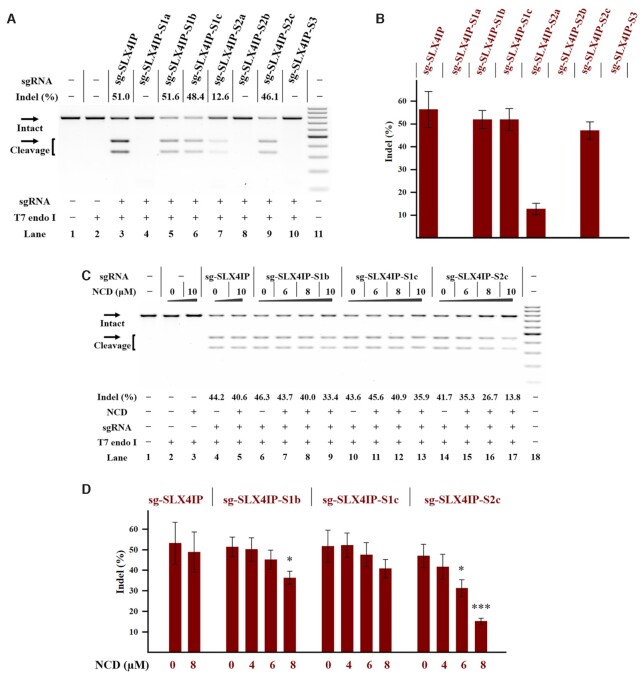
Figure 6.
Ligand control of designer sgRNAs in a stable Cas9-expressing cell line Cellular studies were performed using HeLa-OC cells as described in the Experimental Section. The treatment for each sample is indicated by the signs at the bottom of each lane. All samples were tested in three biological replicates. Image of representative data is shown here. (A) Editing of SLX4IP gene in HeLa-OC cells using the indicated sgRNAs. Lane 1: target control; lane 2: no sgRNA control; lane 3 contains original sg-SLX4IP; lanes 4–10 contain designer sgRNAs harboring different MBL-binding units; lane 11: DNA marker (GeneRuler 100-bp DNA Ladder). (B) The effect of sequence modification on the function of sgRNAs in cells. (C) Ligand control of designer sgRNAs in HeLa-OC cells. Hela-OC cells were exposed to the NCD ligand for 24 h before being harvested for DNA cleaving activity assessments. Lane 1: target control; lanes 2–3: no sgRNA control; lanes 4–5 contain original sg-SLX4IP; lanes 6–9 contain sg-SLX4IP-S1b; lanes 10–13 contain sg-SLX4IP-S1c; lanes 14–17 contain sg-SLX4IP-S2c; lane 18: DNA marker. (D) Bar graph shows the effect of NCD on the function of sgRNAs in HeLa-OC cells. In each group, the indel formation of NCD-treated cells were compared to that of mock-treated cells. P values less than 0.05 are given one asterisk, and P values <0.001 are given three asterisks. For (A) and (C), uncleaved SLX4IP DNA (773 bp) cut to shorter cleavage fragments (441 bp and 332 bp) are demonstrated. For (B) and (D), data represent the mean of three replicates and were shown as mean ± SEM.