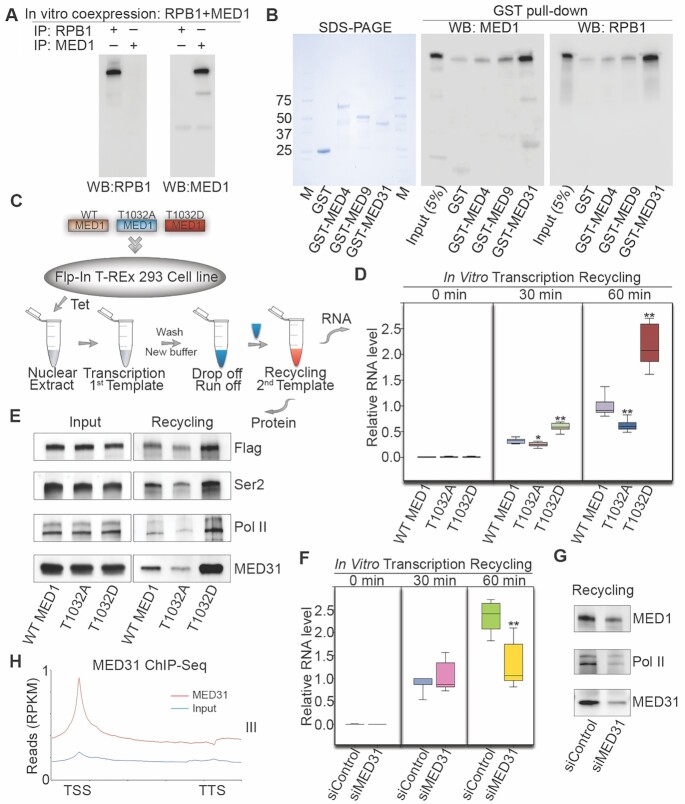
Figure 5.
MED31 bridges pMED1 to Pol II in transcription recycling. (A) Full-length MED1 or RPB1 was expressed using an in vitro cell-free expression system. Purified proteins were incubated overnight and immunoprecipitated with RPB1 or MED1 antibodies, and analyzed by western blot. (B) GST pull-down assays were performed by incubating in vitro translated MED1 or RPB1 with the indicated GST fusion proteins. Western blots were performed using MED1 and RPB1 antibodies. (C) Schematic of transcription recycling assay. Flp-In 293 cell lines were generated stably expressing one single copy of WT or mutated FLAG-MED1. (D) RNA products from the second template reaction were analyzed by reverse transcription followed by qPCR quantification. Regions = 7 (n = 2), P-values were calculated using two-tailed Student’s t-test, *P < 0.01, **P < 0.001. (E) Proteins bound to the second template were assayed by western blots. (F) The transcription recycling assay was performed using nuclear extracts from siControl or siMED31-treated LNCaP-abl cells. RNA products from the second template reaction were analyzed by reverse transcription followed by qPCR quantification. Regions = 7 (n = 2), P-values were calculated using two-tailed Student’s t-test, **P < 0.001. (G) Proteins bound to the second template were assayed by western blotting. (H) Average MED31 ChIP-seq signal densities over the scaled human RefSeq genes of class III in LNCaP-abl cells.