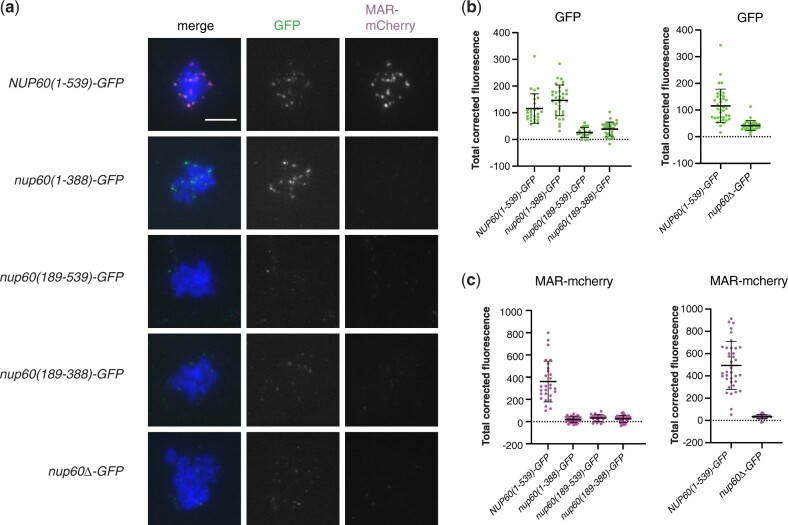
Fig. 7.
Effect of Nup60 truncations on recruitment of the MAR to meiotic chromosomes. a) Representative meiotic chromosome spreads from strains expressing MAR-mCherry and full length or truncated Nup60-GFP fusions. Cells were harvested 10 h after transfer to SPM then spread onto glass slides. The Nup60-GFP fusions were detected using a polyclonal antibody to GFP and an Alexa Fluor 488-conjugated secondary antibody; MAR-mCherry was detected using a polyclonal antibody to mCherry and an Alexa Fluor 594-conjugated secondary antibody. The scale bar in the upper left panel represents 5 µm. All strains are isogenic to the NUP60-GFP strain (SBY6254): nup60(1–388)-GFP (SBY6344), nup60(189–388)-GFP (SBY6341), nup60(189–539)-GFP (SBY6347), and nup60Δ−GFP (SBY6338). b) Quantitation of TCF of Nup60-GFP from spreads represented in (a). Because we see some day-to-day variation in the staining of wild-type spreads, the data are grouped to allow comparison of all samples with a wild-type Nup60-GFP spreads that had been stained on the same day. P = 0.0125, comparing NUP60(1–539) and nup60(1–388) and P < 0.0001 for all other comparisons to (1–539) using one-way ANOVA with Dunnett’s multiple comparisons tests. c) Quantitation of TCF of MAR-mCherry for spreads represented in (a). Data are grouped as in (b). P < 0.0001 for all comparisons to (1–539) using the same method as in (b).