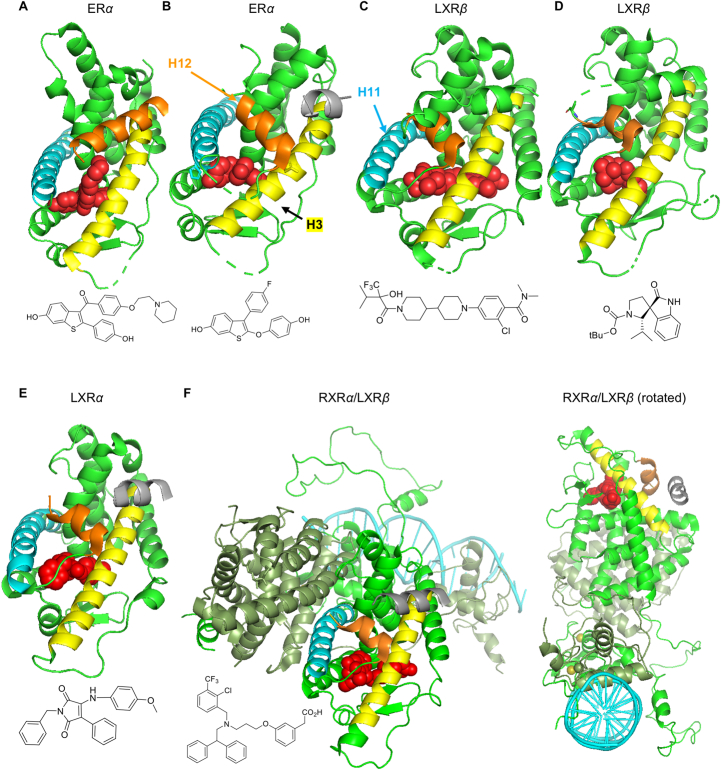
Figure 4.
NHR and ligand structures (H12 orange; H3 yellow; H11 blue; ligand red; coregulator silver). (A) ERα in “antagonist conformation” with SERM raloxifene displacing H12 (PDB 2JFA). (B) ERα in “agonist conformation” with TTC-352 inducing closure, stabilizing H12, and binding the coactivator NCOA2 (PDB 7JHD). (C) Merck Comp9 LXR agonist (LXRβ: PDB 5HJP). (D) LXR “inverse agonist” (LXRβ: PDB 6K9M)375. (E) GSK3986 (LXRα; PDB 2ACL with NCOA1 bound): showing similarity with ER agonist conformation forming AF-2; and similar binding poses with both LXR isoforms and with agonists and inverse agonists. (F) LXRβ:RXRα heterodimer bound to DR-4 DNA element including LBDs, coregulator, and DNA-binding domains (4NQA)376: although containing more components of the transcriptional complex, intrinsically disordered N-terminal domains (containing AF-1) are absent. Tissue selectivity of NR ligands is widely believed to result from cell-specific coregulator expression370; however, the ligand is dominant as the linchpin in allosteric communication between coregulators and DNA377.