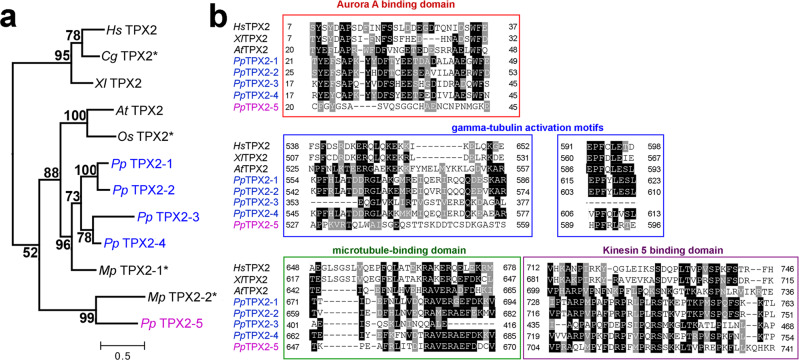
Fig. 1. TPX2 homologs in P. patens.
a Phylogenetic analysis revealed two distinct groups of TPX2 proteins in P. patens: Pp TPX2-1 to −4 (blue), which are more similar to TPX2 genes from seed plants, and atypical TPX2-5 (magenta). Asterisks mark predicted proteins, numbers show bootstrap values. Bar, 0.5 amino acid substitutions per site. Note that AtTPX2L3 and AtTPX2L2 were not added to this tree, since they lack the C-terminal region that is conserved in canonical TPX2 proteins. Hs: Homo sapiens, Gg: Gallus gallus, Xl: Xenopus laevis, At: Arabidopsis thaliana, Os: Oryza sativa, Pp: Physcomitrium patens, Mp: Marchantia polymorpha. b Alignment of TPX2 proteins. Conserved residues are boxed, whereas similar amino acids are hatched.