Fig 4.
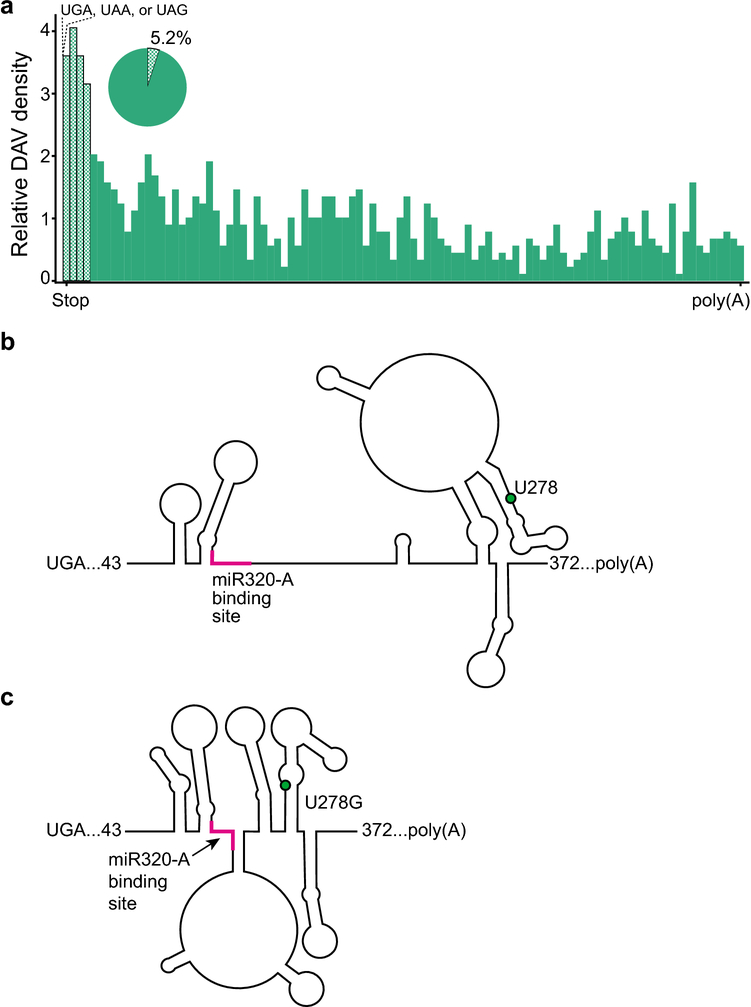
Disease associated variants in the 3’ UTR. a. DAV distribution across 3’ UTRs. The checkered bars indicate potential post-transcriptional regulatory motifs approximately around the ribosomal footprint at the stop codon. Stop codons are labeled above the graph. The pie chart shows proportion of 3’ UTRs that have a DAV within the ribosomal footprint (~15 nucleotides). b. The RNA secondary structure of a section of the FKBP5 3’UTR. The 3’UTR contains the binding site for the miRNA miR320-A (pink). c. The minor allelic SNP rs3800373 changes U to G at position 278 (green dot) causing a structural change that makes the miRNA binding site (pink) less accessible because a greater proportion of the binding site is base paired in a novel stem-loop.