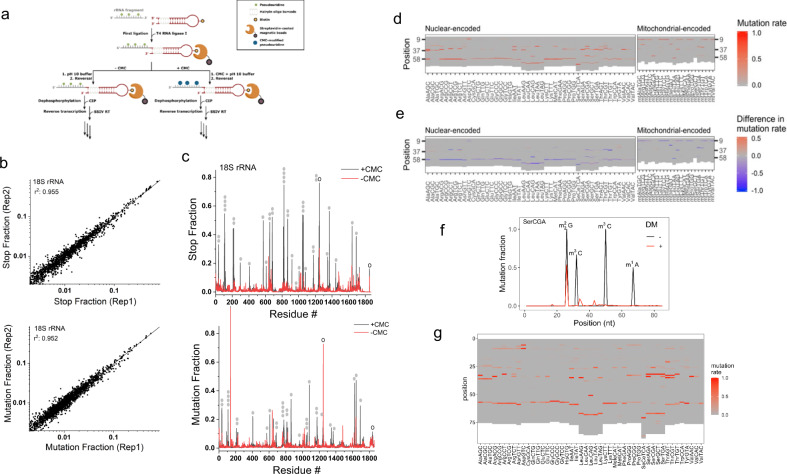
Fig. 2. Sequencing results from chemical and enzymatic treatment on bead.
a Schematic representation of incorporating CMC reaction in MSR-seq for pseudouridine (Ψ) mapping. Total RNA was fragmented, 3′ end-repaired, and ligated to the capture hairpin oligonucleotide. CMC reaction was done on-bead before cDNA synthesis. b CMC reaction mapping replicates: Top graph shows the RT stop fraction and the bottom graph shows the mutation fraction for every residue in human 18 S rRNA among the biological replicates. c Stop and mutation fraction along 18 S rRNA without (red) and with CMC (black) treatment: Top graph shows the RT stop fraction and the bottom graph shows the mutation fraction at each nucleotide position. Known Ψ sites are marked by filled gray ovals, known m1acp3Ψ and m62A sites by open ovals. d Heat map showing the mutation signature at every nucleotide for the most abundant tRNA isodecoder in each isoacceptor family, no demethylase treatment. Nuclear-encoded and mitochondrial-encoded tRNAs are shown separately. e Heat map showing changes in mutation signature upon demethylase (DM) treatment at every nucleotide for the most abundant tRNA isodecoder in each isoacceptor family. Red indicates an increase and blue indicates a decrease in mutation fraction. f Mutation fraction across a tRNASer(CGA) isodecoder ± demethylase treatment (DM) showing the effective removal of base methylation. Known modifications are indicated. g Same as (d), using overnight RT condition for library construction.