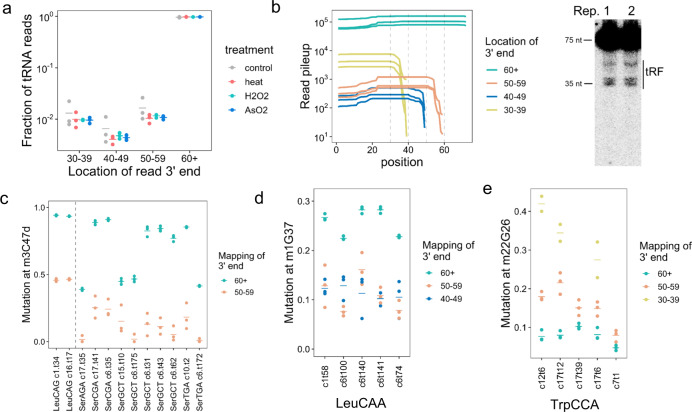
Fig. 6. Analysis of tRNA fragment biogenesis and tRNA modification.
MSR-seq was performed in biological triplicates (n = 3) for each condition. a Total count of tRNA fragment (tRF) relative to full-length tRNA in all four conditions. 60+ tRNA are considered full-length tRNAs; replicates are shown as points, mean of replicates is shown as a bar. b Read pileup of the most abundant tRFGly(CCC) isodecoder showing the amount of tRFs with 3′ ends in 30–39 (anticodon loop), 40–49 (variable loop), and 50–59 (T loop) in unstressed control samples. The gel shows Northern blot tRFGly(CCC) validation. Data for n = 2 independent biological replicates are shown. Source data are provided as a Source Data file. c Comparing mutation fraction of m3C47d in tRF versus full-length cognate tRNA for tRNALeu(CAG) and tRNASer isodecoders in unstressed controls. d Comparing mutation fraction of m1G37 in tRF versus full-length cognate tRNA for tRNALeu(CAA) isodecoders in unstressed controls. e Comparing mutation fraction of m22G26 in tRF versus full-length cognate tRNA for tRNATrp isodecoders in unstressed controls.