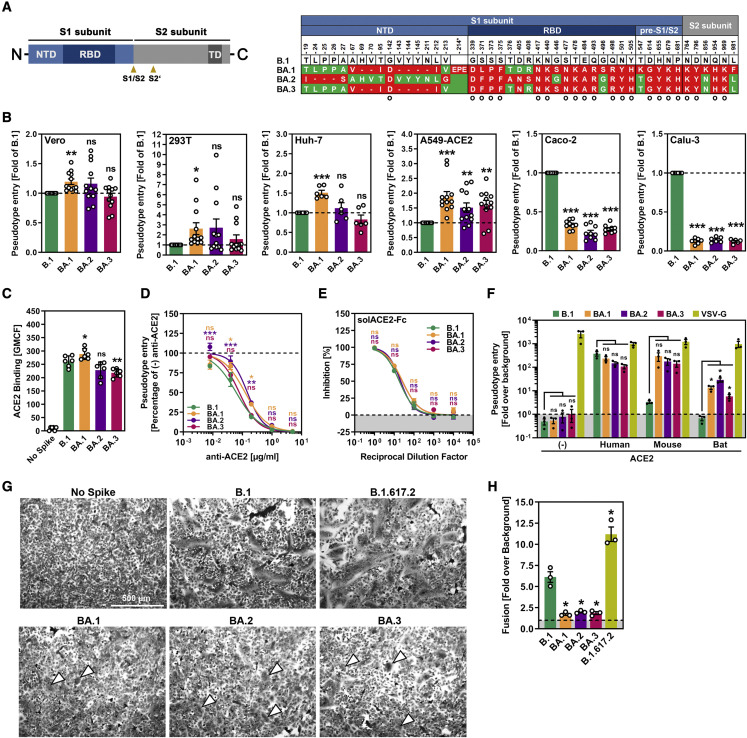
Figure 1.
S proteins of Omicron sublineages do not exhibit major differences in ACE2 usage or ability to drive cell-cell and virus-cell fusion
(A) Schematic overview of the SARS-CoV-2 spike (S) protein domain structure (left) and summary of the mutations found in the different Omicron sublineages (right; numbering is according to the S protein of SARS-CoV-2 B.1). S protein residues that are identical between the S proteins of some Omicron sublineages and B.1 are marked in green, whereas mutated residues are highlighted in red (note: the BA.1 S protein harbors an insertion between amino acid residues 214 and 215). Further, mutations found in all Omicron sublineages are indicated by a circle. Abbreviations: NTD, N-terminal domain; RBD, receptor-binding domain; TD, transmembrane domain; S1/S2 and S2’, cleavage sites in the S protein.
(B) S-protein-driven cell entry. We added particles bearing the indicated S proteins (or no S protein) to the indicated cell lines and analyzed cell entry by measuring the activity of virus-encoded firefly luciferase in cell lysates 16–18 h after inoculation. Presented are the average (mean) data from 6–12 biological replicates (each conducted with four technical replicates) in which cell entry was normalized against B.1 (set as 1). Error bars show the SEM. Statistical significance was assessed by two-tailed Student’s t tests (p > 0.05, not significant [ns]; p ≤ 0.05, ∗; p ≤ 0.01, ∗∗; p ≤ 0.001, ∗∗∗). Please also see Figure S1.
(C) ACE2 binding efficiency. 293T cells transiently expressing the indicated S proteins (or no S protein) where first incubated with soluble ACE2 fused to the Fc portion of human immunoglobulin G (solACE2-Fc) and subsequently incubated with an Fc-specific AlexaFluor-488-coupled secondary antibody; then, solACE2-Fc binding was analyzed by flow cytometry. Presented are the average (mean) data from six biological replicates (each conducted with single samples) in which ACE2 binding was normalized against B.1 (set as 1). Error bars show the SEM. Statistical significance was assessed by two-tailed Student’s t tests (p > 0.05, not significant [ns]; p ≤ 0.05, ∗; p ≤ 0.01, ∗∗).
(D) Blockade of S-protein-driven cell entry by an anti-ACE2 antibody. Vero cells were preincubated with serial dilutions of anti-ACE2 antibody before particles bearing the indicated S proteins were added. S-protein-driven cell entry was analyzed and normalized to samples without anti-ACE2 antibody (set as 1). Presented are the average (mean) data of three biological replicates, each performed with four technical replicates. Error bars show the SEM. Statistical significance was assessed by two-way analysis of variance with Dunnett’s post hoc tests (p > 0.05, not significant [ns]; p ≤ 0.05, ∗; p ≤ 0.01, ∗∗; p ≤ 0.001, ∗∗∗). Please also see Tables S1 and S2.
(E) Blockade of S-protein-driven cell entry by solACE2-Fc. Particles bearing the indicated S proteins were preincubated with serial dilutions of solACE2-Fc (or no solACE2-Fc) before being added to Vero cells. S-protein-driven cell entry was analyzed and normalized to samples without solACE2-Fc (= 0% inhibition). Presented are the average (mean) data of three biological replicates, each performed with four technical replicates. Error bars show the SEM. Statistical significance was assessed by two-way analysis of variance with Dunnett’s post hoc tests (p > 0.05, not significant [ns]).
(F) Usage of mouse and bat ACE2 by Omicron S proteins. BHK-21 cells transiently expressing human, mouse, or horseshoe bat (Rhinolophus pearsonii) ACE2 orthologs (or no ACE2) were inoculated with particles bearing the indicated S proteins or VSV glycoprotein (VSV-G). Cell entry of pseudovirus particles was analyzed and normalized to particles bearing no viral glycoprotein (set as 1; indicated by dashed line). Presented are the average (mean) data of three biological replicates, each performed with four technical replicates. Error bars show the SEM. Statistical significance was assessed by two-tailed Student’s t tests (p > 0.05, not significant [ns]; p ≤ 0.05, ∗).
(G) Qualitative fusion assay. A549-ACE2 cells transfected to express the indicated S proteins (or no S protein) were fixed 24 h after transfection and stained with May-Gruenwald and Giemsa solution before microscopic images were taken (scale bar, 500 μm). Arrowheads indicate small syncytia in cells expressing the S proteins of the different Omicron sublineages.
(H) Quantitative fusion assay. 24 h after transfection, 293T cells transiently expressing the indicated S proteins (or no S protein) along with the beta-galactosidase alpha fragment were resuspended and seeded on top of A549-ACE2 cells transiently expressing the beta-galactosidase omega fragment. After an additional 24 h of incubation, beta-galactosidase substrate was added, and luminescence was recorded. Presented are the average (mean) data of three biological replicates, each performed with four technical replicates. Error bars show the SEM. Statistical significance was assessed by two-tailed Student’s t tests (p ≤ 0.05, ∗).