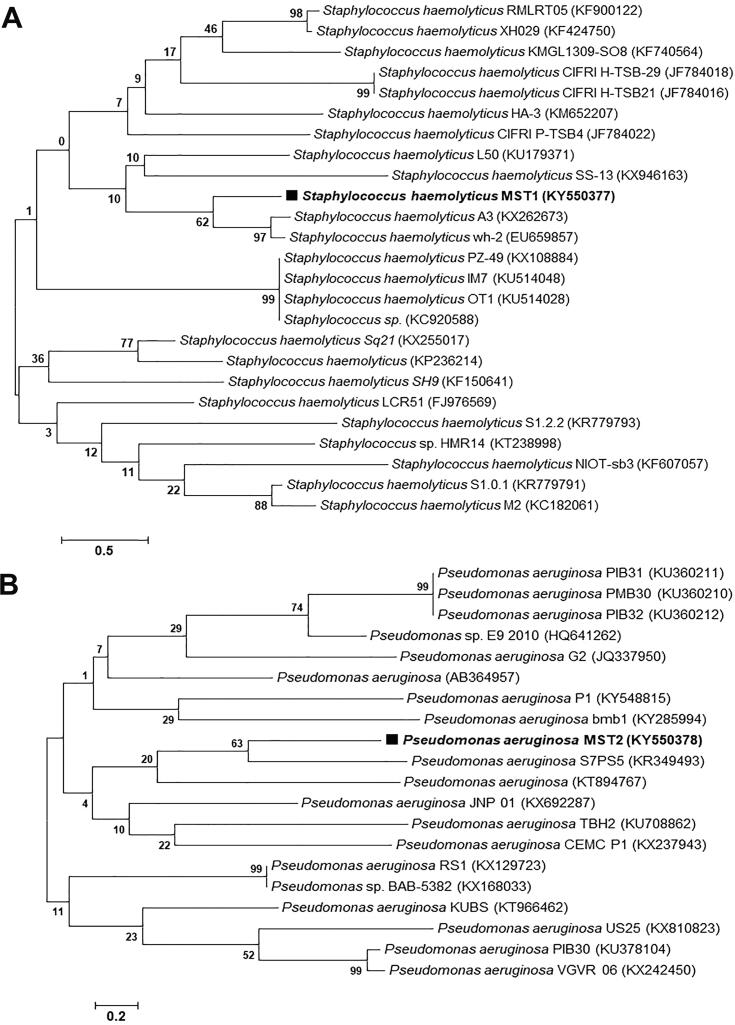
Fig. 4.
Phylogenetic trees of (A) S. haemolyticus MST1 (KY550377) and (B) P. aeruginosa MST2 (KY550378) reveal their evolutionary relationships with regards to the closest bacteria on the basis of 16S rRNA nucleotide sequences retrieved from the GenBank database, and accession numbers of the sequences are indicated in parentheses. The phylogenetic trees were constructed employing the Neighbor-Joining tree with bootstrap values of 500 replicates, and the bars denote Jukes-Cantor distances of 0.5 and 0.2 for S. haemolyticus MST1 and P. aeruginosa MST2, respectively.