Figure 5.
Evolutionary and expression analyses of NAC genes.
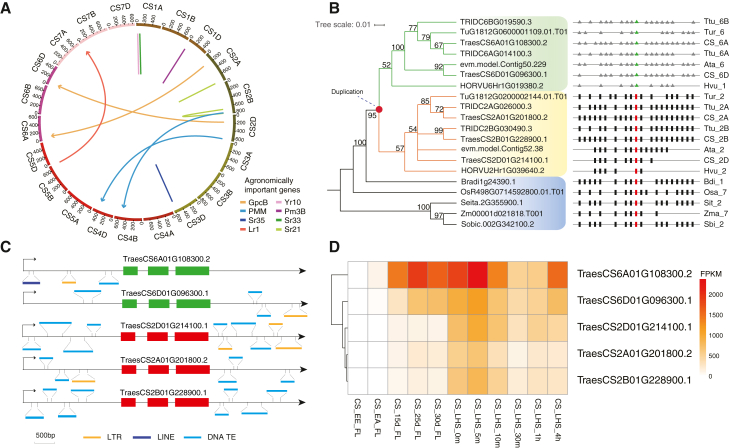
(A) Circos plot showing eight previously identified important genes that have experienced the RBGD. The known agronomically important genes are associated with stem rust resistance (Sr21, Sr33, Sr35), stripe rust resistance (Yr10), leaf rust resistance (Lr1), powdery mildew resistance (Pm3B), phosphomannomutase (PMM), and earlier senescence and higher grain protein, iron, and zinc content (GPC). The arrow/line represents the direction of gene duplication from the ancestral gene to the newly duplicated copy.
(B) Maximum likelihood phylogeny of the NAC genes and the syntenic regions that contain NAC genes in other Poaceae genomes. A red solid circle in the phylogenetic tree represents one of the RBGD duplication events that created a duplicated copy in chromosome 6 of the common ancestor of Triticeae species. The right side of the phylogenetic tree presents the syntenic regions with NAC genes. The identified syntenic relationships among genes shown as black and red rectangles suggest that the genes in group I are positionally conserved, and therefore the ancestral copies, whereas the genes in group II that are illustrated as green triangles surrounded by gray triangles are the new, duplicated copies.
(C) Schematic diagrams showing the gene structure and flanking TEs around NAC genes in CS. Different types of TEs are indicated by bars with different colors.
(D) Expression levels of NAC genes in CS. The duplicated copy of TraesCS6A01G108300 has the highest expression in the flag leaf among the five NAC genes. EE, ear emergence; EA, anthesis; LHS, leaf under heat stress.