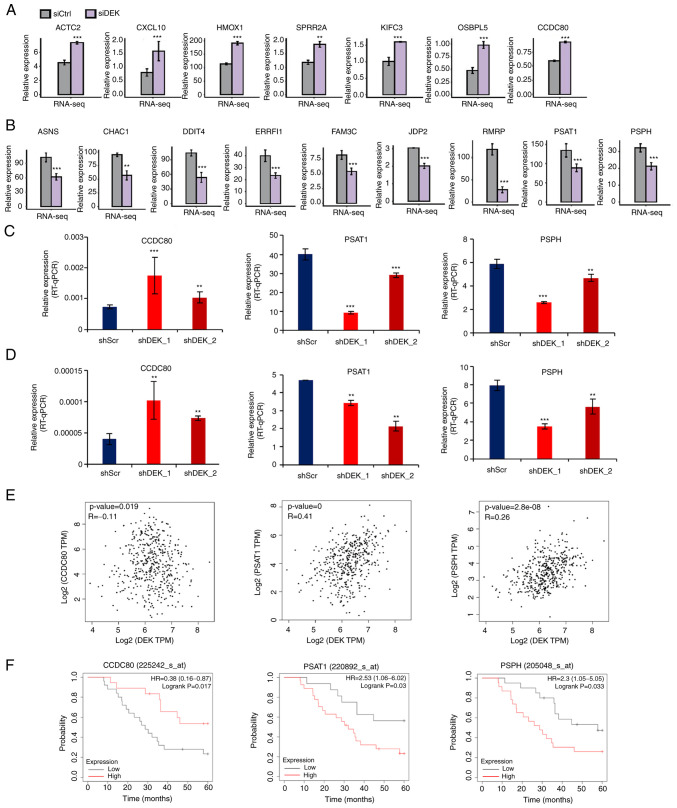
Figure 3.
Representative genes whose expression is potentially regulated by DEK. (A and B) FPKM values of representative DEGs that were significantly upregulated or downregulated in the siDEK condition in a human GC cell line AGS are shown as bar plots. Gray and purple bars show the FPKM values of the siCtrl- and siDEK-treated AGS cells, respectively. Bars and error bars represent means and SEMs, respectively. **P<0.01, ***P<0.001. (C and D) Relative expression levels of DEGs (% of GAPDH) in shRNA-infected cells in AGS and NUGC4 cell lines were validated by RT-qPCR assay, respectively. Statistical differences between groups (shScr and either shDEK_1 or shDEK_2) were tested using one-way ANOVA with Newman-Keuls multiple comparisons test, and P<0.05 was considered statistically significant. Bars and error bars represent the mean ± SD, respectively (n=3). **P<0.01, ***P<0.001. (E) Correlation of gene expression levels between DEK and CCDC80, PSAT1 and PSPH from public datasets are shown as dot plots. Each dot represents a single GC case. All the data were derived from the GEPIA2 database (http://gepia2.cancer-pku.cn/#correlation). (F) Kaplan-Meier curves of GC patients as stratified with high and low expression levels of CCDC80, PSAT1 and PSPH. All of the data were obtained from the GSE22377 (N=43) dataset. (http://kmplot.com/analysis/index.php?p=service&cancer=gastric). P-values were calculated by log-rank test. GC, gastric cancer; FPKM, fragments per kilobase of transcript per million fragments mapped; DEGs, differentially expressed genes; ACTA2, alpha smooth muscle actin; CXCL10, chemokine (C-X-C motif) ligand 10; HMOX1, heme oxygenase-1; SPRR2A, small proline-rich protein 2A; KIFC3, kinesin family member C3; OSBPL5, oxysterol binding protein like 5; CCDC80, coiled-coil domain containing 80; ASNS, asparagine synthetase; CHAC1, cation transport regulator 1; DDIT4, DNA damage-inducible transcript 4; ERRFI1, ERBB receptor feedback inhibitor 1; FAM3C, FAM3 metabolism regulating signaling molecule C; JDP2, Jun dimerization protein 2; RMRP, RNA component of mitochondrial RNA processing; PSAT1, phosphoserine aminotransferase 1; PSPH, phosphoserine phosphatase.