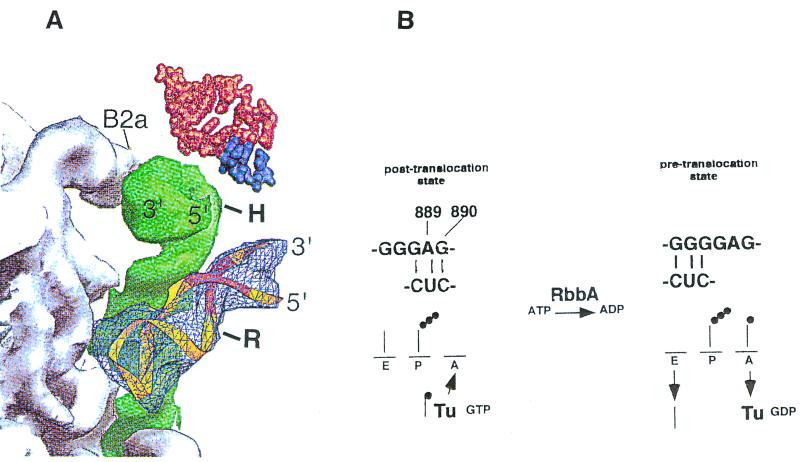
FIG. 6.
Switch helix in the 30S subunit. A view of the packing interactions from the platform side of the A-site tRNA is shown. (A) Model for the action of hygromycin B and of RbbA on 70S ribosomes. The 889-890 base pair switch helix in the 30S subunit is shown. The diagram (from reference 7) shows the penultimate stem (green) between the 50S subunit (gray) and the switch helix (bp 900) (light blue), and the remainder of the 30S subunit with A-site tRNA (red) and mRNA (dark purple) above. B2a, interface contact, shown for orientation; H, approximate location of base 1408, which is protected by hygromycin B from dimethyl sulfate modification; R, site of binding of RbbA to base G925 of the switch helix region. A ribbon representation of the 889-890 stem-loop region of 16S rRNA containing the S turn is in yellow, with the switch helix region in orange. (B) Proposed model for RbbA. The conformational switch in 16S rRNA demonstrated by Lodmell and Dahlberg is at the top (22). RbbA enhances modification by diethyl pyrocarbonate of A889 and RNase T1 cleavage of G890 (21), suggesting a switch to the conformation on the right that increases binding to the A site. The conformation on the left restricts binding to the A site. RbbA-mediated hydrolysis of ATP may be an energy requirement for the transition. The transition may also be mediated by the EF-Tu–GTP-dependent binding of aminoacyl-tRNA to the A site, since RbbA can bind EF-Tu. The binding of hygromycin B to bases 1494 to 1408 could affect the bp 900 switch helix, preventing the binding of RbbA to this region.