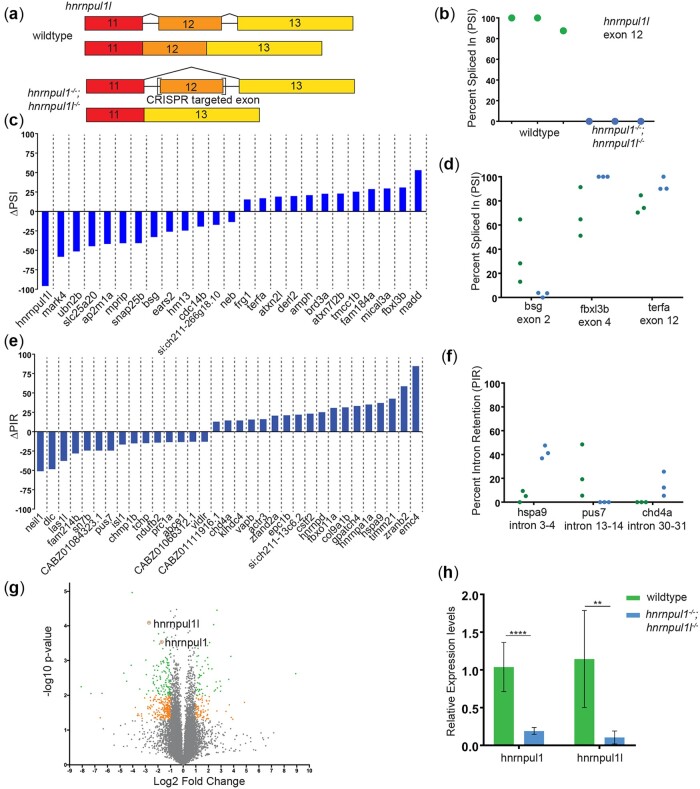
Fig. 2.
Loss of hnrnpul1l and hnrnpul1 leads to differential splicing. a) Schematic showing the processing of hnrnpul1l to form standard transcript and AS of exon 12 of the hnrnpul1l gene as a result of CRISPR-Cas9 targeted mutagenesis. b) PSI for exon 12 of hnrnpul1l in wild type and hnrnpul1/1l mutant embryos at 3 dpf. c) Change in PSI of all exon skipping events in hnrnpul1/1l mutant embryos compared to wild types. d) Detailed view of PSI of genes associated with phenotypes, points represent each biological replicate. e) Change in percent intron retention (PIR) of all intron retention events in hnrnpul1/1l mutant embryos compared to wild types. f) Detailed view of PIR of genes associated with phenotypes, points represent each biological replicate. Details of affected exon/intron are given in Supplementary Table 1. g) Volcano plot showing all differentially expressed genes. Gray points = 1>Log2 FC>-1 or P > 0.05, orange points = 1<Log2 FC<-1 and P ≤ 0.05, green points = 1<Log2 FC<-1 and P ≤ 0.01. h) qPCR validation hnrnpul1 and hnrnpul1l expression in hnrnpul1/1l mutants at 3 dpf compared to wild types. **P ≤ 0.01, ****P ≤ 0.0001, determined by Student’s t-test.