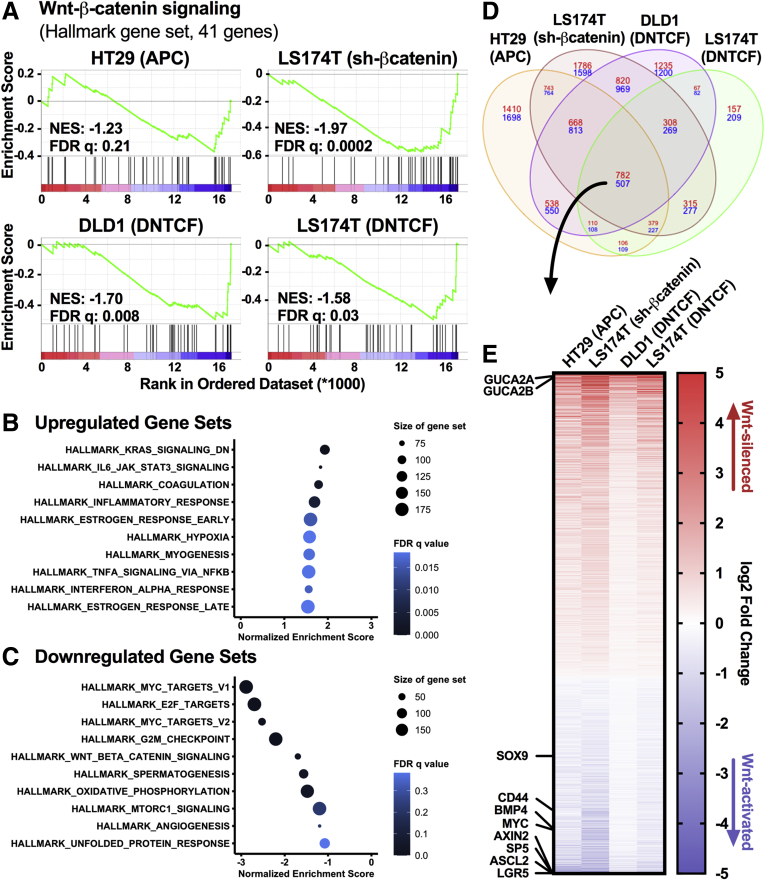
Figure 2.
The GUCY2C ligands are among the most sensitive targets of β-catenin/TCF signaling. HT29(APC) cells were treated with 300 μM zinc for 24 hours, LS174T(shβ-catenin) cells were treated with 1 μg/mL DOX for 72 hours, DLD1(DNTCF) cells were treated with 1 μg/mL DOX for 24 hours, and LS174T(DNTCF) cells were treated with 1 μg/mL DOX for 48 hours. RNA-seq and differential gene expression analysis was performed in cells treated with or without the respective inducing agent. Unbiased gene set enrichment analysis on each cell line compared the ranked log fold change of 17,229 genes to the 50 hallmark gene sets maintained by the Molecular Signatures Database (MSigDB). (A) The Wnt-β-catenin signaling gene set enrichment plot for each cell line, with the corresponding normalized enrichment score and false discovery rate (FDR) q value. (B, C) The top 10 upregulated and downregulated gene sets for DLD1(DNTCF) cells, ranked by normalized enrichment score. Color indicates FDR q value, and size of the data points indicates the number of genes in each gene set. (D) Differential gene expression in the 4 cell lines by RNA-seq reveals 782 upregulated (red) and 507 downregulated (blue) genes upon silencing Wnt signaling. (E) Heatmap of the 1289 differentially expressed genes ranked by log2 fold change. GUCA2A and GUCA2B are the 7th and 12th most upregulated transcripts. RNA-seq results represent the average of 3 replicates.