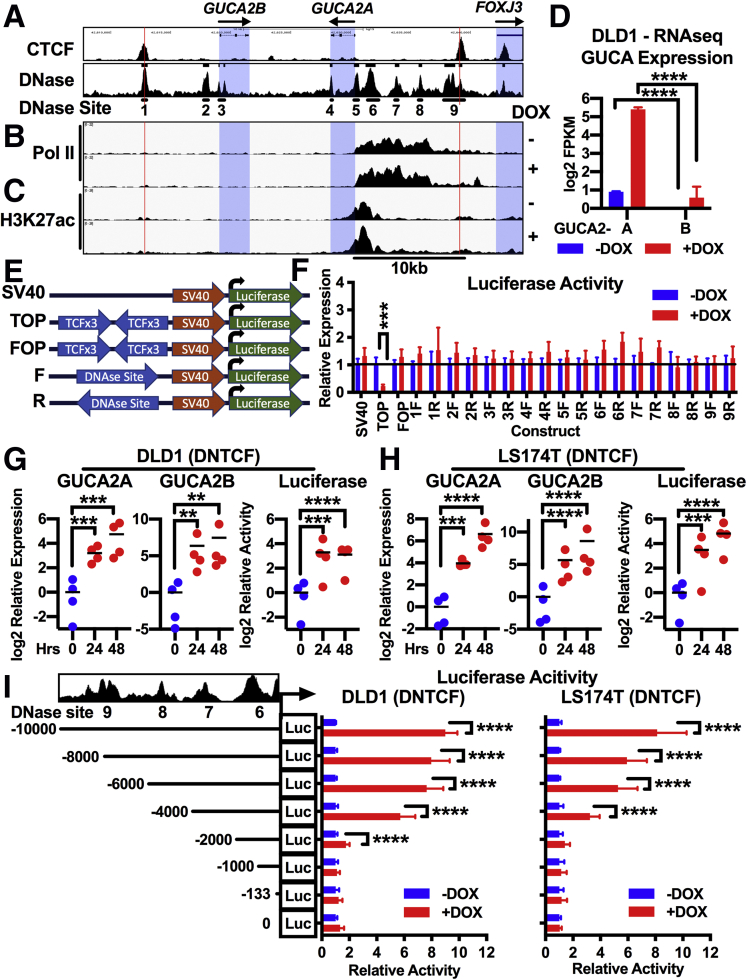
Figure 6.
(See previous page). A Pol II-rich super-enhancer upstream of GUCA2A confers Wnt sensitivity. (A) Nine regions of DNase hypersensitivity were identified from sequencing tracks in Figure 4: CTCF ChIP-seq from human HCT116 colon cancer cells (GSE92879) and DNase-seq from transverse colon (GSE90398; hg19, chr1:42,607,745–42,644,356). ChIP-seq of (B) Pol II and (C) H3K27ac in DLD1(DNTCF) cells with (+) or without (–) 1 μg/mL DOX for 24 hours. (D) Corresponding RNA-seq FPKM quantification illustrating greater transcript expression of GUCA2A than GUCA2B in DLD1(DNTCF) cells. (E, F) Nine regions of DNase hypersensitivity were selected and cloned into luciferase reporters. (E) Diagram of enhancer-luciferase reporters driven by a constitutive (SV40) promoter with no upstream enhancer, upstream TCF sites (TOP), upstream mutant TCF sites (FOP), or upstream GUCA2A DNase-sensitive sites in forward (5′-3′) or reverse (3′-5′) orientation. (F) Luciferase constructs were expressed in DLD1(DNTCF) cells, and luciferase was quantified with (+) or without (–) 1 μg/mL DOX for 24 hours. (G, H) A luciferase construct driven by the region from +15 to –10,000 relative to the GUCA2A transcription start site was expressed in (G) DLD1(DNTCF) and (H) LS174T (DNTCF) cells. GUCA2A mRNA, GUCA2B mRNA, and luciferase activity were quantified following 1 μg/mL DOX for 0–48 hours. (I) Luciferase constructs containing 5′ truncations of the region from +15 to –10,000 relative to the GUCA2A transcription start site were expressed in DLD1(DNTCF) and LS174T (DNTCF) cells and luciferase activity was quantified following 1 μg/mL DOX for 24 hours (DLD1) or 48 hours (LS174T). (B, C) ChIP-seq tracks represent the average of 2 replicate immunoprecipitations. (D) RNA-seq results represent the average of 3 replicates. (G, H) Data points represent the average of 3 wells of cells from a single experiment, with the mean of 4 independent experiments indicated. (F, I) Bars represent the average of 4 independent experiments ± SEM. Significance was determined by (G, H) 1-way, or (F, I) 2-way analysis of variance with matched analysis for independent experiments on log2-transformed results. Data are presented relative to noninduced cells. ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.