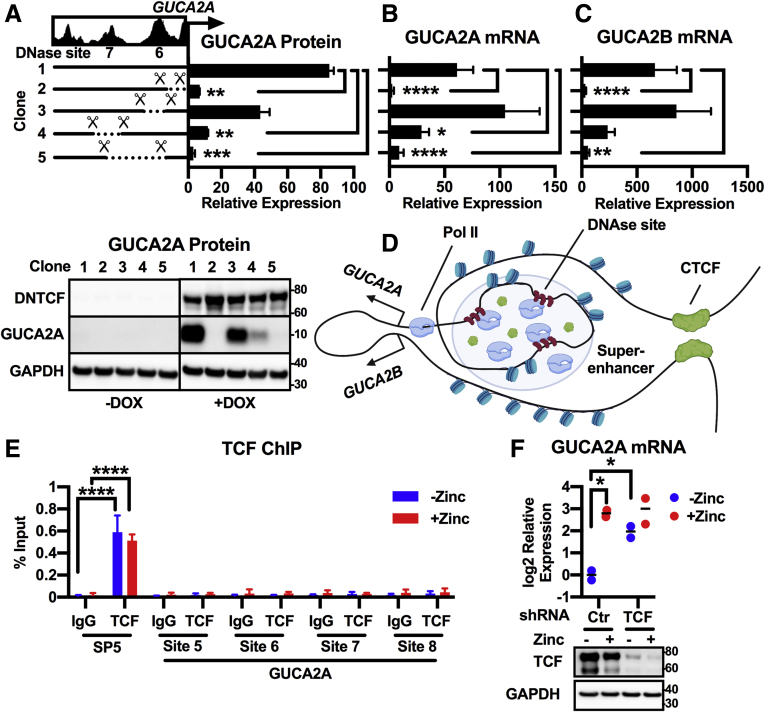
Figure 8.
Cas9 deletion confirms a super-enhancer necessary for β-catenin-TCF-sensitive locus control. DLD1(DNTCF) cells expressing Cas9 and untargeted (clone #1) or GUCA2A-locus-targeted gRNAs harbor biallelic deletions encompassing the GUCA2A promoter (clone #2), DNase site #6 (clone #3), DNase site #7 (clone #4), or both DNase sites (clone #5). Expression of (A) GUCA2A protein, (B) GUCA2A mRNA, and (C) GUCA2B mRNA was quantified with (+) or without (–) 1 μg/mL DOX for 24 hours. (D) Hypothetical model of GUCA2A and GUCA2B promoter positioning relative to a Pol II-rich super-enhancer region upstream of GUCA2A, consisting of multiple DNase sites. (E) ChIP-pcr in HT29(APC) cells treated with (+) or without (–) 300 μM zinc for 24 hours reveals enrichment of TCF at the promoter of a Wnt target gene, SP5, with a TCF-specific antibody, but not with control IgG. In contrast, TCF was not detected at sites within 6kb of the GUCA2A TSS: DNase site #5 (GUCA2A promoter), site #6, site #7, or site #8. (F) GUCA2A mRNA expression in HT29(APC) cells stably expressing an untargeted (Ctr) or TCF-targeted shRNA, and treated with (+) or without (–) 300 μM zinc for 24 hours. GUCA2A mRNA expression is retained despite TCF knockdown, illustrated by Western blot. (A–C) Bars represent the average of (A) 2 or (B, C) 3 independent experiments ± SEM, and data are presented relative to noninduced cells. Significance was determined by 2-way analysis of variance with matched analysis for independent experiments on log2-transformed results. (E) Bars represent the mean ± SD of 3 IPs, and data are presented relative to input DNA. Significance was determined by 2-way analysis of variance. (F) Data points represent the average of 3 wells of cells from a single experiment, with the mean of 2 independent experiments indicated. Significance was determined by 2-way analysis of variance with matched analysis for independent experiments on log2-transformed results. Data are presented relative to noninduced cells receiving control shRNA. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.