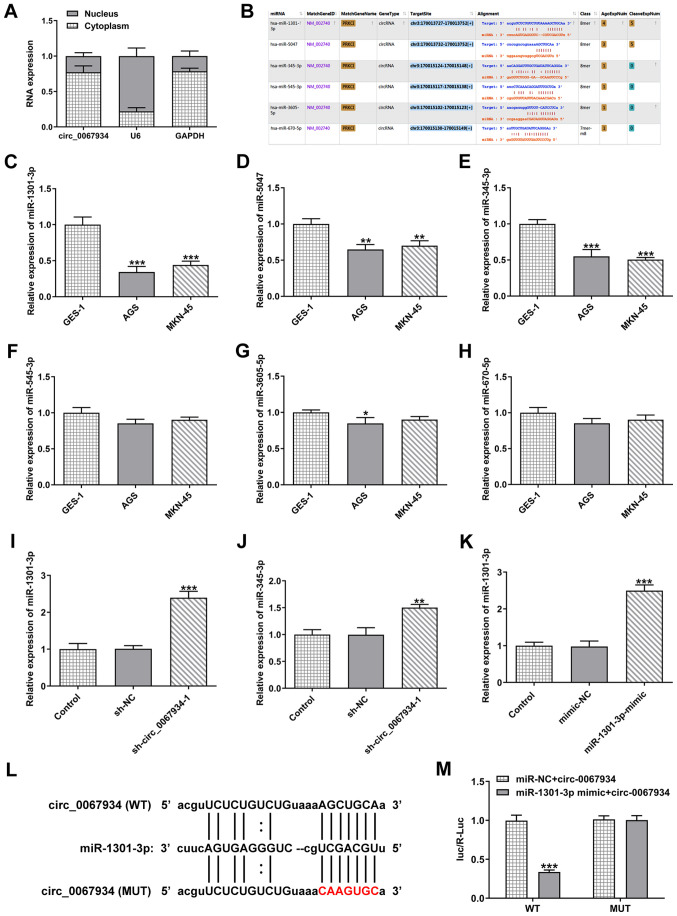
Figure 3.
circ_0067934 targets miR-1301-3p. (A) Expression level of circ_0067934 in the cytoplasm and nucleus of AGS cells was detected via RT-qPCR. (B) The starBase database was used to predict the binding site of circ_0067934 with miR-5047, miR-1301-3p, miR-670-5p, miR-345-3p, miR-545-3p and miR-3605-5p. The expression levels of (C) miR-1301-3p, (D) miR-5047, (E) miR-345-3p, (F) miR-545-3p, (G) miR-3605-5p and (H) miR-670-5p in gastric cancer cell lines was identified via RT-qPCR. The expression levels of (I) miR-1301-3p and (J) miR-345-3p were detected after transfection with sh-circ_0067934. (K) miR-1301-3p mimics increased miR-1301-3p expression in AGS cells. (L) The predictive binding sequence between circ-0067934 and miR-1301-3p. The red letters in the circ-0067934 showed the mutated sites. (M) Luciferase activity was determined using a luciferase reporter assay. Data are expressed as the mean ± SD. *P<0.05, **P<0.01, ***P<0.001 vs. GES-1 cells, sh-NC or mimic-NC. RT-qPCR, reverse transcription-quantitative PCR; circ, circular RNA; sh, short hairpin RNA; NC, negative control; miR, microRNA; WT, wild-type; MUT, mutant.