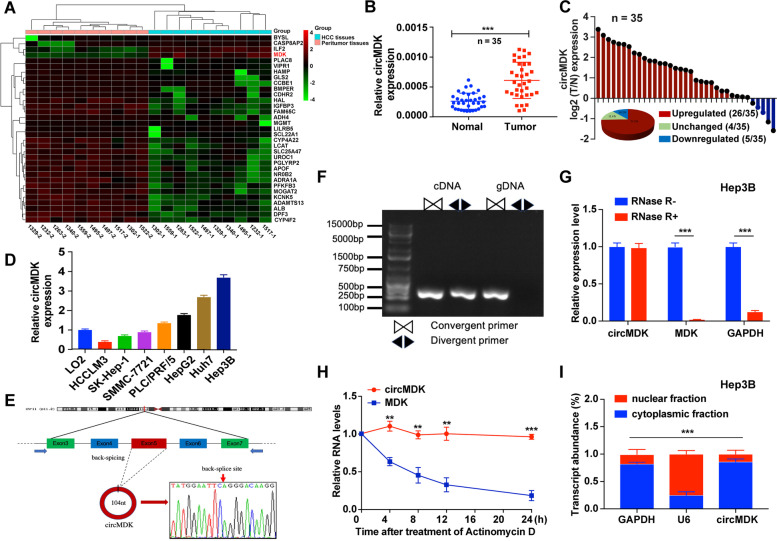
Fig. 1.
CircMDK expression profiling reveals that circMDK is upregulated in HCC. A Clustered heat map of the 42 dysregulated circRNAs in HCC tumor and peritumor tissues. The red and green strips represented high and low expression, respectively. B Analysis for RNA levels of circMDK in 35 paired samples of HCC were determined by qRT-PCR. The error bars represent standard deviation (SD) (n = 35). ***p < 0.001. C Histogram and pie chart of the proportions of HCC samples in which circMDK expression was upregulated (26/35, 74.3%, red), downregulated (4/35, 14.2%, blue), or no change (5/35, 11.4%, green), respectively. Log2 (T/N expression) value > 1 as significantly higher expression, < -1 as lower expression, and between -1 and 1 as no significant change. T, tumorous tissue; N, nontumorous tissue. D Analysis for RNA levels of circMDK in HCC cell lines (HCCLM3, SK-Hep-1, SMMC-7721, PLC/PRF/5, HepG2, Huh7 and Hep3B) and normal liver cell line (LO2). E The back-splice junction site of circMDK was identified by Sanger sequencing. F PCR analysis for circMDK and MDK in complementary DNA (cDNA) and genomic DNA (gDNA). G Analysis for RNA levels of circMDK and MDK after treated with RNase R in Hep3B cells. ***p < 0.001. H qRT-PCR for the abundance of circMDK and MDK mRNA in Huh7 cells treated with Actinomycin D at the indicated time points. **p < 0.01; ***p < 0.001. I Levels of circMDK in the nuclear and cytoplasmic fractions of Hep3B cells. ***p < 0.001. Data are shown as mean ± SD of three independent experiments