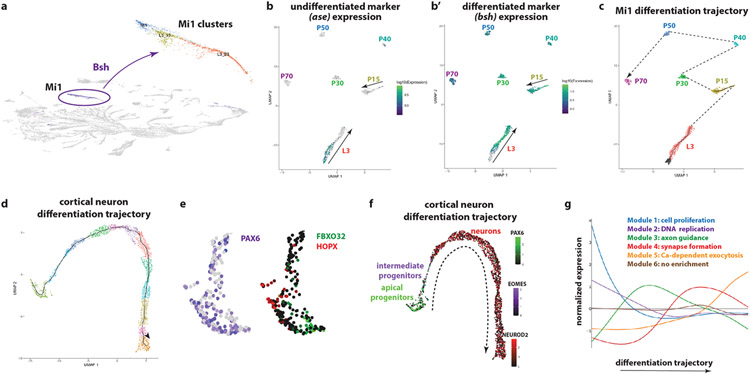
Extended Data figure 9: Neuronal differentiation in flies and humans.
(a) Bsh is expressed almost exclusively in Mi1s and was used to identify the Mi1 clusters. Cluster Mi1 represents the pupal annotated cluster. Cluster L3_23 consists of GMCs that give rise to Mi1s and newly born Mi1s, while cluster L3_53 corresponds to more mature Mi1 cells, as assessed by their proximity to the P15 Mi1 cells.
(b) UMAP plot of Mi1 cells at different stages of differentiation from L3 to P70. The expression of ase (b) and bsh (b’) were used to find the beginning and end, respectively, of the L3 trajectory.
(c) UMAP plot of Mi1 cells at different stages of differentiation from L3 to P70. L3 and P15 trajectories are elongated, depicting Mi1 cells of different ages. Transcriptomes are then synchronized (compact group of clusters) around P30.
(d) UMAP plot showing the trajectory of 3,363 single cell transcriptomes of the developing human cortex (gestational week 19), as generated by Monocle3. The orientation of the trajectory was identified by looking at the expression of marker genes for progenitors, intermediate progenitors and neurons (see panel f). Colors indicate different clusters along the trajectory.
(e) UMAP plot focusing on the PAX6-positive single-cell transcriptomes of the developing human cortex (gestational week 19). The radial glia population contains both ventricular radial glia (FBXO32-positive cells) and outer radial glia (HOPX-positive cells).
(f) UMAP plot of 3,363 single-cell transcriptomes of the developing human cortex (gestational week 19). The trajectory from progenitors to neurons can be observed by the expression of Pax6 (apical progenitors), Eomes (intermediate progenitors), and NeuroD2 (neurons). The dashed arrow depicts the differentiation trajectory.
(g) Differential expression analysis along the trajectory of the cortical neurons identified six modules of genes. Gene Ontology enrichment analysis found the first two modules to be enriched in terms such as cell proliferation (FDR=10−3) and DNA replication (FDR=10−44); they likely correspond to the progenitor cells. Then, the third module is enriched in neurite development terms, such as axon guidance (FDR=10−7), while the fourth one is enriched in terms related with synapse organization (FDR=10−11). The fifth one contains “functional genes”, such as calcium-dependent exocytosis (FDR=10−2). The sixth module does not show a clear peak of expression and no GO terms were found to be enriched.