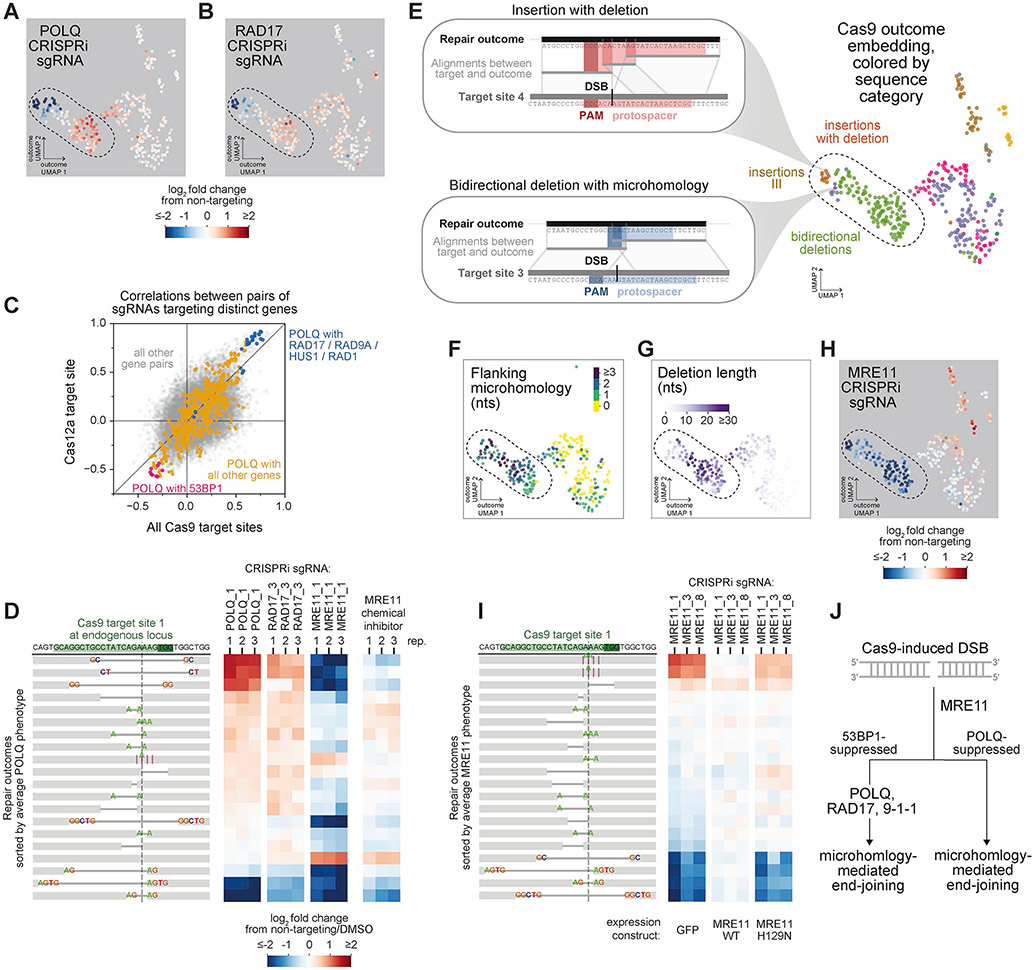
Figure 6. Repair-seq delineates microhomology-mediated end-joining pathways.
(A+B) Log2 fold changes in outcome frequencies produced by indicated CRISPRi sgRNAs overlaid on the UMAP embedding of Cas9 outcomes. See also Figure 3G.
(C) Comparison of correlations between outcome redistribution signatures for pairs of sgRNAs targeting distinct genes between composite Cas9 screens and Cas12a screen.
(D) Effects of indicated CRISPRi sgRNAs or of chemical MRE11 inhibitor on repair outcome frequencies at Cas9 target site 1 in the endogenous HBB locus. Heatmaps display log2 fold changes in outcome frequency for the 20 outcomes with highest baseline frequency, sorted by average log2 fold change across POLQ sgRNA replicates.
(E) Examples of Cas9-induced mutations consistent with known Pol θ-mediated architectures (insets). Top black lines represent the sequence of a repair outcome; bottom grey line represents the sequence flanking the targeted DSB in the integrated screening vector; lines between these depict local alignments; and red (top) and blue (bottom) rectangles mark the protospacer and PAM of Cas9 target sites.
(F+G) Deletions in UMAP embedding of Cas9 outcomes colored by the length of microhomology flanking the deletion junction (F) or the length of sequence removed by the deletion (G).
(H) Log2 fold changes in outcome frequencies produced by an MRE11-targeting sgRNA overlaid on the UMAP embedding of Cas9 outcomes. The region marked with a dotted line includes the vast majority of MRE11-promoted outcomes and is similarly indicated in panels (A, B, E, F, and G).
(I) Effects of indicated CRISPRi sgRNAs on outcome frequencies in cells ectopically expressing MRE11 + GFP, or nuclease-inactive MRE11 (H129N) + GFP, or control (GFP). Heatmaps (right) display log2 fold changes in the frequencies of the 20 outcomes with highest baseline frequency, sorted by average log2 fold change across MRE11-targeting sgRNAs in control cells.
(J) Model of genetically distinct sub-pathways of resection-initiated end joining.