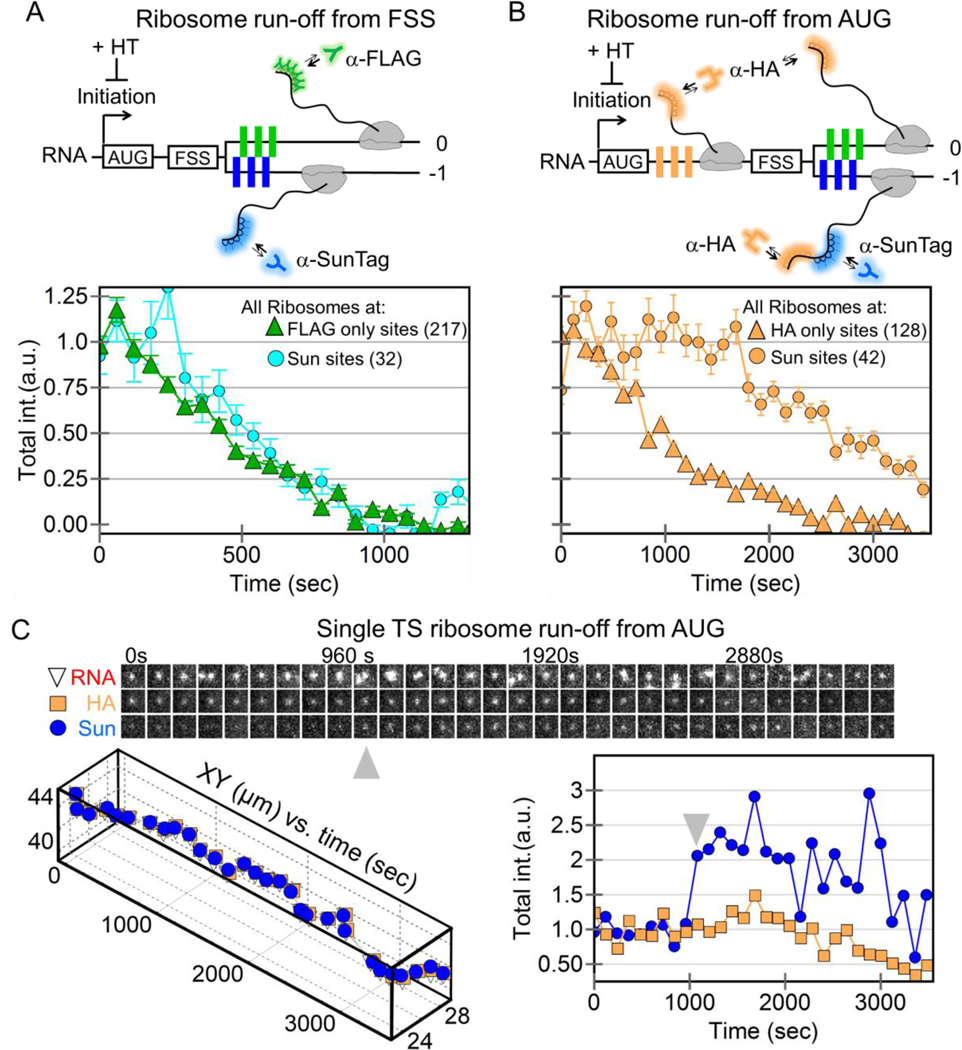
Figure 4. Ribosomal run-off at frameshifting and non-frameshifting translation sites.
(A) A schematic showing harringtonine-induced ribosomal run-off from the +FSS 2x multi-frame RNA reporter with FLAG (green) and SunTag epitopes (blue) in the 0 and −1 frames. The intensity of nascent chain signals within non-frameshifting translation sites (marked by just anti-FLAG Fab, green triangles, 217 translation sites initially in 19 cells) and frameshifting sites (marked by anti-SunTag scFv, cyan circles, 32 translation sites initially) drops with time as ribosomes complete translation, i.e. run-off. (B) Same as A, but with a modified multi-frame reporter with extra HA epitopes (orange, HA multi-frame tag) upstream of the frameshift sequence. The intensity of nascent chain signals from non-frameshifting translation sites (marked by just anti-HA Fab, orange triangles, 175 translation sites initially in 27 cells) and frameshifting translation sites (marked by anti-SunTag scFv, orange circles, 42 translation sites initially) drops with time as ribosomes run-off. (C) A sample single translation site encoding the modified HA multi-frame reporter (shown in B) after addition of harringtonine. A montage of image trims shows the detected RNA-, HA-, and Sun-signals through time. Below, the positions of the detected signals within the site are plotted through time. On the right, the normalized total intensity of the HA Fab signal (marking all ribosomes) and the SunTag scFv signal (marking frameshifting ribosomes) is plotted through time. Gray arrows signify a burst of frameshifting, coinciding with multi-RNA interactions.