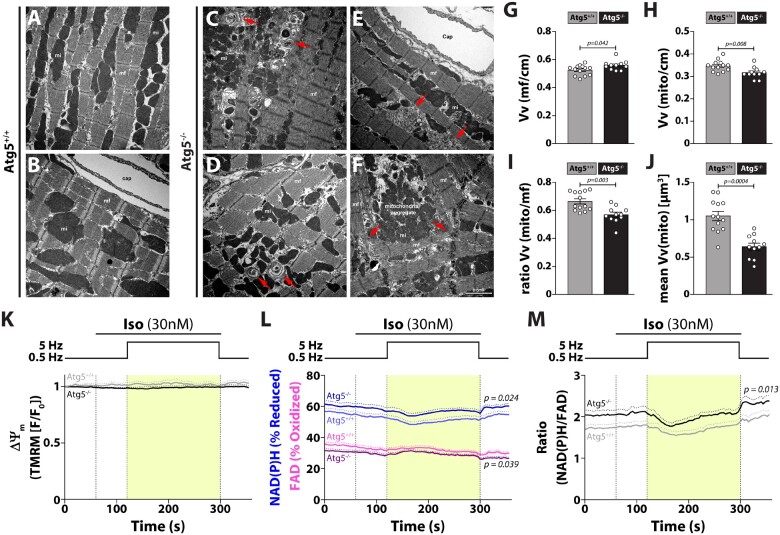
Figure 6.
Atg5-deficiency causes aberrant cardiomyocyte composition, decreased mitochondrial abundance and functional overcompensation of individual mitochondria. (A–F) Representative transmission electron micrographs of longitudinal sections of cardiomyocytes depict ultrastructural changes due to the absence of basal autophagy. Note onion-like membrane aggregation (red arrows in C and D), membrane cisternae (red arrows in E), sarcomere disarray, misalignment of myofibrils (red arrows in F), and aggregation of mitochondria (F). Scale bar, 2 µm; Cap, capillary; mf, myofibril; mi, mitochondria; cm, cardiomyocytes. Relative volume of (G) myofibrils Vv(mf/cm) and, (H) mitochondria Vv(mito/cm) per cardiomyocyte from Atg5+/+ and Atg5−/− mouse hearts. (I) Ratio between the relative volume of mitochondria and myofibrils, Vv(mito/cm)/Vv(mf/cm). (J) Volume-weighted mean volume of mitochondria, meanVv(mi), was used to quantify mitochondrial volume and size distribution in Atg5+/+ vs. Atg5−/− mouse hearts. (G–J) Data show mean ± SEM of N = 11–13 mice per group, indicated P-values were calculated using Mann–Whitney test comparing Atg5−/− to the Atg5+/+ control. Isolated Atg5−/− and Atg5+/+ cardiomyocytes were exposed to the stress protocol as in Figure 3A. (K) Mitochondrial membrane potential (ΔΨm; determined by TMRM fluorescence normalized to resting fluorescence, F/F0), (L) NAD(P)H/NAD(P)+ and FADH2/FAD (autofluorescence determined in the same cells) and, (M) ratio of reduced NAD(P)H to oxidized FAD (data from L) were determined. (K–M) Data show mean ± SEM, n = 28/18 cells from 3/2 Atg5+/+/Atg5−/− mice, respectively, for TMRM measurements, and n = 33/32 cells from 3/3 Atg5+/+/Atg5−/− mice, respectively, for NAD(P)H/NAD(P)+ and FADH2/FAD measurements. P-values were calculated using Bonferroni’s post hoc test, following significant two-way repeated-measures ANOVA.