Figure 1: Overview of experimental design.
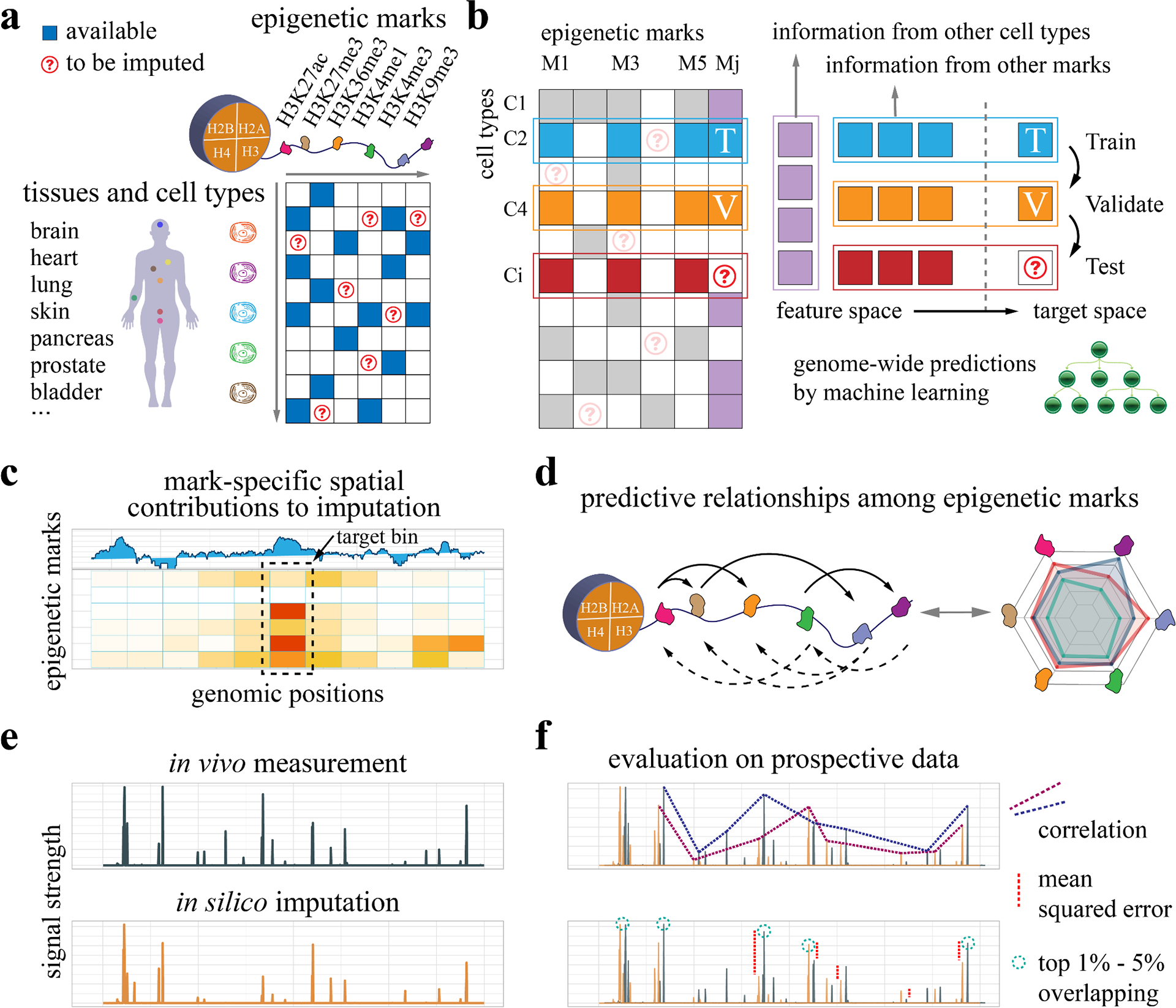
a, We develop machine learning models to impute genome-wide epigenetic signal tracks across cell types and investigate the regulatory relationships among epigenetic marks based on available data. b, For each cell type-mark combination to be imputed, we consider both the information from other cell types of the same mark and the information from other marks in the same cell type. The available data are partitioned into the training and validation sets to build machine learning models for epigenome imputation. c, To investigate the cross-prediction relationships among epigenetic marks, we dissect the machine learning models and extract the spatial contribution of each feature epigenetic mark to a target mark. d, The pairwise modulatory relationships among marks are summarized. The relationships are directional and asymmetric, where the solid and dashed arrows represent predicting others and being predicted respectively. e, The in silico imputation from our approach is compared with held-out in vivo measurement, which is collected prospectively to avoid information leakage or overfitting. f, The imputed data are compared with observed data based on multiple evaluation metrics, including correlations, mean squared errors (MSEs), and overlapping for the top 1% - 5% regions.
