Figure 7. The neuron‐glial antigen 2 (NG2) tamoxifen‐inducible Cre recombinase (CreERTM) driver specifically labels mural cells, whereas the inducible platelet‐derived growth factor receptor (PDGFR)βCreERT2 line lacks specificity, also labeling a significant fraction of PDGFRα+ cardiac fibroblasts.
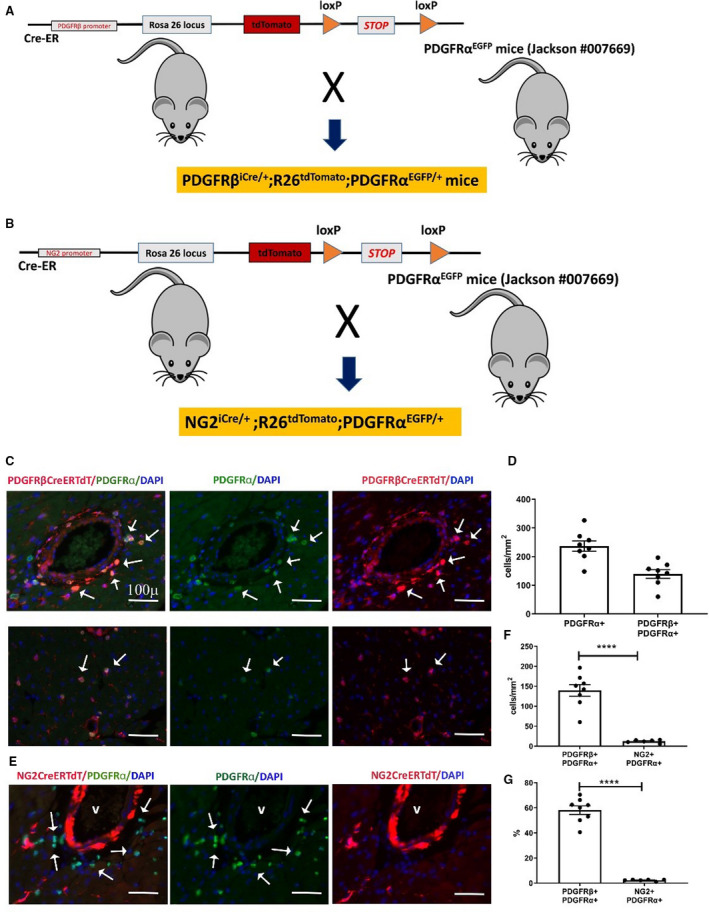
A and B, To test the specificity of the inducible PDGFRβ and NG2‐Cre drivers for myocardial pericytes, we bred PDGRβiCre/+;R26tdTomato mice (A), and NG2iCre/+;R26tdTomato animals (B) with fibroblast reporter PDGFRαEGFP mice. Schematic cartoons show the breeding scheme. C, Dual immunofluorescence of left ventricular sections shows that the inducible PDGFRβ‐Cre driver lacks specificity, labeling a large fraction of perivascular fibroblasts (C, top row) and interstitial fibroblasts (C, lower row) in the myocardium. PDGFRβ+/PDGFRα+ fibroblasts are indicated with arrows. D, Quantitative analysis (n=8) shows the density of PDGFRα+ fibroblasts and PDGFRα+/PDGFRβ+ fibroblasts in the myocardium. E, In NG2iCre/+;R26tdTomato;PDGFRαEGFP mice, staining for tandem dimer Tomato (tdTom) and green fluorescent protein shows that cardiac fibroblasts are not labeled for NG2, supporting the specificity of the inducible NG2‐Cre line. F, Quantitative analysis shows that the number of PDGFRβ+ cells identified as fibroblasts is markedly higher than the number of NG2+ cells that exhibit PDGFRα expression (****P<0.0001; n=6–8). G, Approximately 60% of PDGFRα+ fibroblasts are also labeled for PDGFRβ, whereas the percentage of fibroblasts labeled for NG2 is low (****P<0.0001; n=6–8). Scalebar=100 μm. Data are expressed as mean±SE. Statistical comparisons were performed using unpaired t test. DAPI indicates 4’,6’‐diamidino‐2‐phenylindole; and EGFP, enhanced green fluorescent protein.
