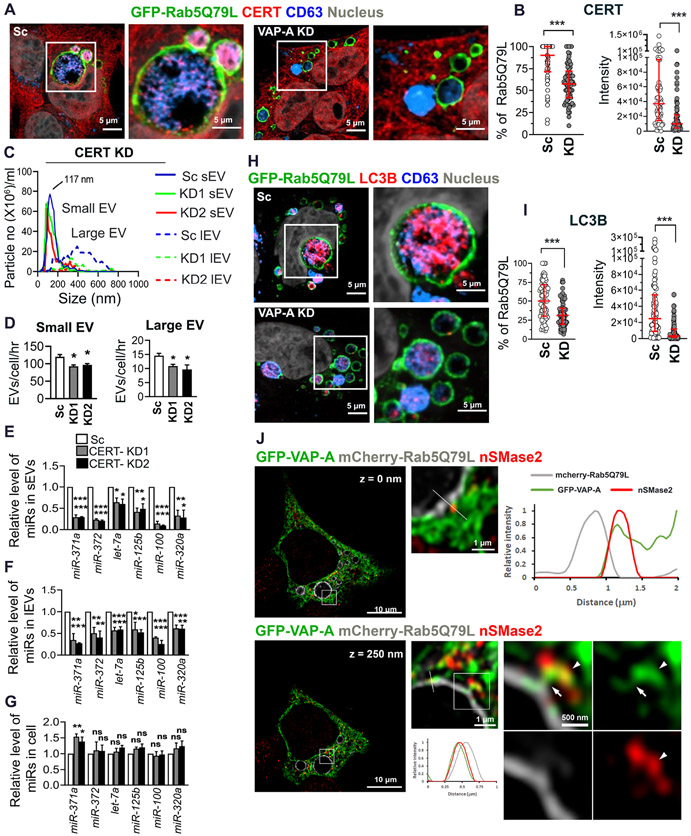
Figure 7: CERT controls the number and RNA content of EVs.
(A) Representative merged images of GFP-Rab5Q79L-transfected DKs-8 cells (scrambled control (Sc) and VAP-A-knockdown (KD)) with CERT and CD63 immunofluorescence staining. Selected areas are enlarged at the right.
(B) Quantitation of the percentage of CERT-positive GFP-Rab5Q79L rings and the intensity of CERT presented in GFP-Rab5Q79L rings. Each circle represents a cell. n = 68 and 70 cells from 3 independent experiments for Sc and KD, respectively.
(C) Graph shows NTA traces of small and large EVs purified from control (Sc) and CERT-KD (KD1, KD2) DKs-8 cells. Median values were plotted from three independent experiments.
(D) Small and large EV release rates from control and CERT-KD cells calculated from three independent NTA datasets.
(E-G) qRT PCR analysis of miRNA levels in control and CERT-KD small and large EVs, and their parental cells, normalized to U6 snRNA. Data from three independent experiments.
(H) Representative merged images of GFP-Rab5Q79L-transfected DKs-8 cells (scrambled control (Sc) and VAP-A-knockdown (KD)) with LC3 and CD63 immunofluorescence staining. Selected areas are enlarged at the right.
(I) Quantitation of the percentage of LC3-positive GFP-Rab5Q79L rings and the intensity of LC3 presented in GFP-Rab5Q79L rings. Each circle represents a cell. n = 75 and 69 cells from 3 independent experiments for Sc and KD, respectively.
(J) Representative deconvolved and merged images of GFP-VAP-A and mCherry-Rab5Q79L-co-transfected DKs-8 cell with nSMase2 immunofluorescence staining. Selected areas are enlarged at the right. Lines were scanned for the intensity of each fluorescence channel of the images and plotted at the right for 0 nm and at the bottom for 250 nm Z-step images. Arrows indicate a bridge of GFP-VAP-A between the limiting membrane of an mCherry-Rab5Q79L ring and nSMase2. Arrowheads indicate nSMase2 association with the ER.
Data plotted as mean +/− S.E.M for bar graphs and as median and interquartile range for scatter plots. *p<0.05, **p<0.01, ***p<0.001. ns, not significant.