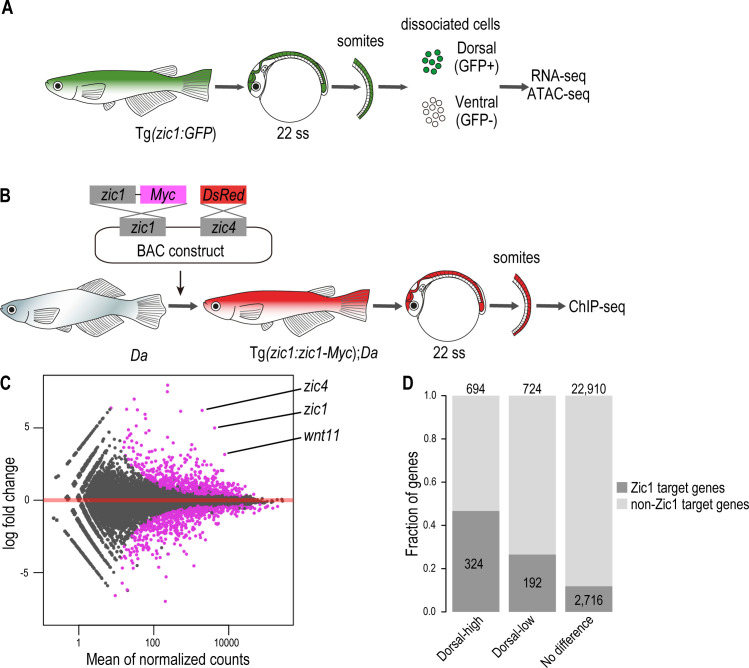
Figure 5. Zic1 regulates dorsal-specific expression of genes in the somites.
(A) Schematic representation of preparation of dorsal and ventral somite cells for RNA-seq and ATAC-seq. (B) Schematic representation of generating the transgenic line Tg(zic1:zic1-Myc);Da. Somites expressing zic1-Myc are subjected to ChIPmentation against Myc. (C) Analysis of RNA-seq revealed that 694 genes are expressed specifically in the dorsal somites (among these genes are zic1, zic4 and wnt11) and 724 genes are expressed specifically in ventral somites. Magenta indicates differentially expressed genes (adjusted p-value < 0.01). (D) The ChIPmentation against Zic1 revealed that 324 of dorsal-high genes, 192 dorsal-low genes and 2716 non differential expressed genes in the somites are potential Zic1 target genes.