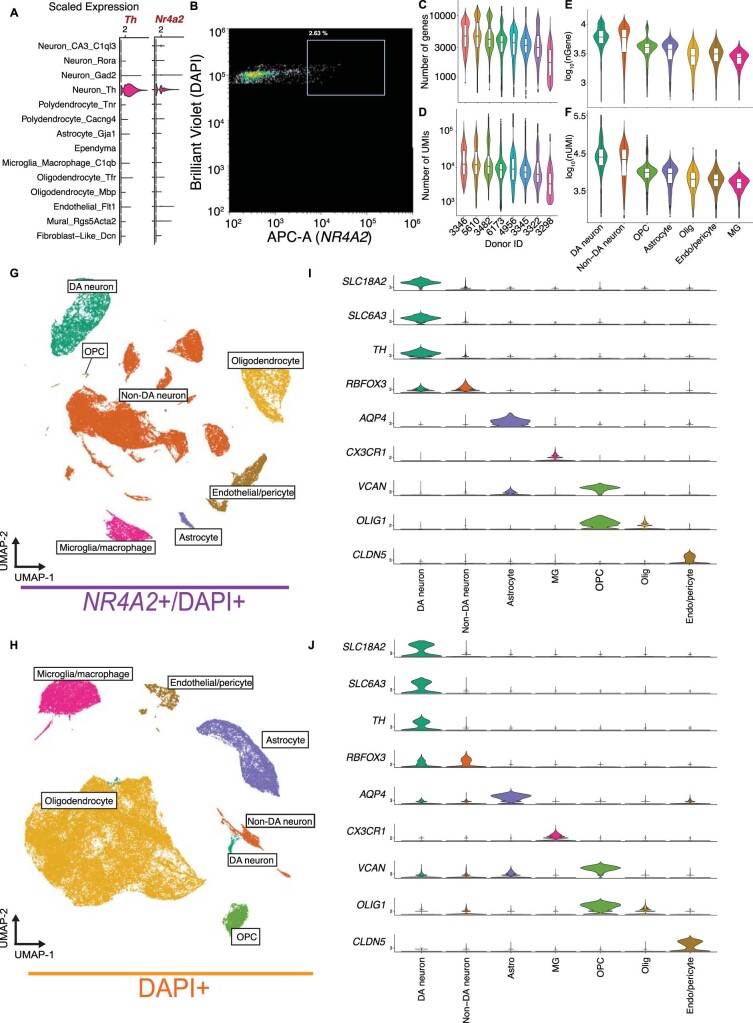
Extended Data Fig. 1. High-throughput snRNA-seq profiling of SNpc.
a, Expression of tyrosine hydroxylase (Th) and Nr4a2 in a published scRNA-seq study of mouse midbrain(21). b, Representative FANS plot for enriching midbrain dopaminergic neurons. APC-A (x-axis) represents NR4A2 antibody channel and Brilliant Violet (y-axis) represents DAPI channel. The NR4A2 gate was thresholded to select the top 2.5–4% of all nuclei (red box). c,d, Number of genes (C) and UMIs (unique molecular indicators) (D) per nucleus grouped by donor (n = 11,577 nuclei for subject 3298; 49,759 nuclei from 3322; 29,230 nuclei for 3345; 23,747 nuclei for subject 3346; 24,856 nuclei for subject 3482; 25,243 nuclei for subject 4956; 6,967 for subject 5610; and 13,294 nuclei for subject 6173). e-f, log10 number of genes (nGene) (E) and log10 number of UMIs (nUMI) (F) per cell type (n = 15,684 DA neuronal nuclei; 46,860 non-DA neurons; 5,866 OPC nuclei; 14,579 astrocyte nuclei; 76,837 oligodendrocyte nuclei; 16,755 microglia/macrophage nuclei; and 8,092 endothelial cell/fibroblast nuclei). For all box plots, center bar indicates median value and lower and upper hinges correspond to first and third quartiles respectively. Whisker distance from upper and lower hinges represents ≤1.5× !QR. DA, dopaminergic; non-DA, non-dopaminergic, OPC, oligodendrocyte precursor cell; Olig, oligodendrocyte; Endo/pericyte, endothelial cells/pericytes; MG, microglia/macrophage. g,h, UMAP representations of the NR4A2 + (G) and NR4A2- (H) gated nuclei, colored by cluster. i,j, Violin plots of marker genes for the seven major cell classes in the NR4A2 + (I) and NR4A2- (J) gated nuclei. DA, dopaminergic; non-DA, non-dopaminergic; Astro, astrocyte; MG, microglia/macrophages; OPC, oligodendrocyte precursor cell; Olig, oligodendrocyte; Endo, endothelial cells.