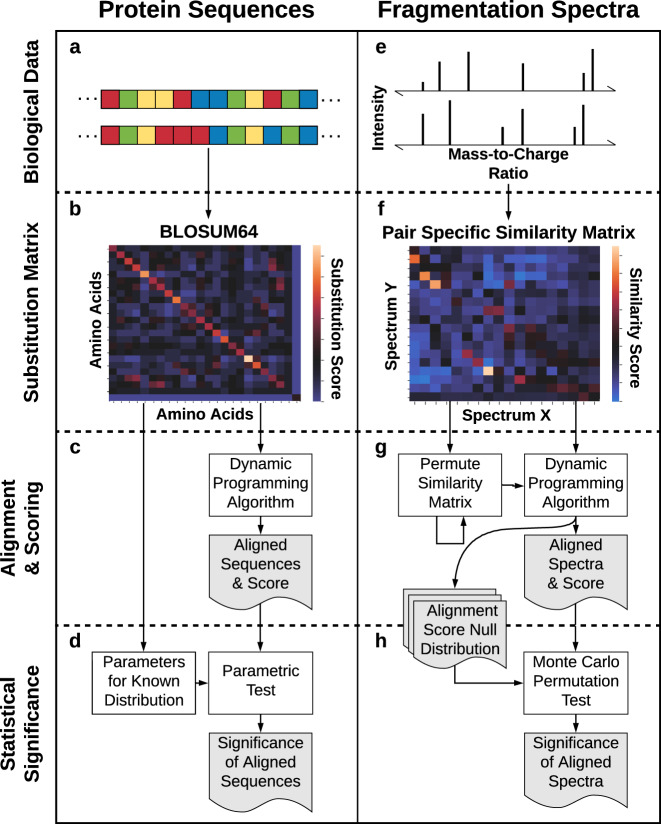
Fig. 1. Analogous to how protein sequences undergo alignment, SIMILE aligns fragmentation spectra with allowances for substitutions and gaps.
a–c Pairs of protein sequences are globally aligned by using a dynamic programming algorithm such as Needleman–Wunsch with an evolutionary model-based substitution matrix and gap penalty. d Significance of the alignment is calculated by assuming alignment scores that follow a distribution parameterized by choice of substitution matrix. e–g Likewise, pairs of fragmentation spectra are aligned by a dynamic programming algorithm with a pair-specific similarity matrix and a gap penalty of zero. g, h The alignment-score null distribution used to calculate the significance of observed alignments stems from permuting pair-specific similarity matrix interrelating fragmentation spectra X and Y randomly many times with restrictions according to the null hypothesis.