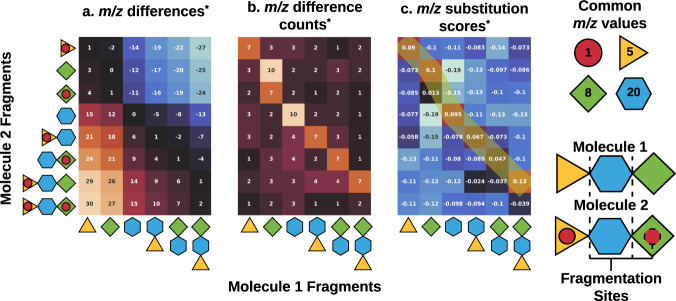
Fig. 2. For two hypothetical molecules, this cartoon illustrates the underlying steps of the SIMILE algorithm in calculating pairwise m/z similarity matrices.
For this example, two molecules “1” and “2” are shown. Each molecule is made from three blocks and there are two modifiers to molecule 2. a All pairwise m/z differences between fragments of molecule 1 and molecule 2 are stored in a matrix. b All entries with the same m/z difference in the top-two quadrants are replaced by the number of entries in which that m/z difference occurred. The same process is repeated for the bottom-two quadrants. c Calculating the pseudoinverse of the directed graph laplacian with this matrix yields similarity scores for each pair of m/z values. The quadrants interrelating molecules 1 and 2 can then be fed into a dynamic programming algorithm to yield aligned fragment ions between molecules 1 and 2. For illustrative purposes, only the quadrant corresponding to molecule 1 vs. molecule 2 of the full matrices is shown. The other quadrants corresponding to molecule 1 vs. molecule 1, molecule 2 vs. molecule 1, and molecule 2 vs. molecule 2 are still used for calculations.