Abstract
Most ectotherms obey the temperature-size rule, meaning they grow larger in a colder environment. This raises the question of how the interplay between genes and temperature affects the body size of ectotherms. Despite the growing body of literature on the physiological life-history and molecular genetic mechanism underlying the temperature-size rule, the overall genetic architecture orchestrating this complex phenotype is not yet fully understood. One approach to identify genetic regulators of complex phenotypes is quantitative trait locus (QTL) mapping. Here, we explore the genetic architecture of body-size phenotypes, and plasticity of body-size phenotypes at different temperatures using Caenorhabditis elegans as a model ectotherm. We used 40 recombinant inbred lines (RILs) derived from N2 and CB4856, which were reared at four different temperatures (16, 20, 24, and 26 °C) and measured at two developmental stages (L4 and adult). The animals were measured for body length, width at vulva, body volume, length/width ratio, and seven other body-size traits. The genetically diverse RILs varied in their body-size phenotypes with heritabilities ranging from 0.0 to 0.99. We detected 18 QTL underlying the body-size traits across all treatment combinations, with the majority clustering on Chromosome X. We hypothesize that the Chromosome X QTL could result from a known pleiotropic regulator—npr-1—known to affect the body size of C. elegans through behavioral changes. We also found five plasticity QTL of body-size traits where three colocalized with body-size QTL. In conclusion, our findings shed more light on multiple loci affecting body-size plasticity and the possibility of co-regulation of traits and traits plasticity by the same loci under different environments.
Subject terms: Genetic mapping, Quantitative trait, Genetic linkage study, Ecophysiology
Introduction
The body sizes of ectotherms such as invertebrates, insects, and fish are negatively correlated with their ambient temperature, where warmer environments result in smaller body size. Besides body sizes, the ectotherms’ life-history traits are also strongly affected by temperature. Phenotypic plasticity (the phenotypes that can be expressed by a single genotype at different environmental conditions) due to temperature changes has been studied widely for many different ectotherms, including evolutionary, ecological, physiological, and molecular investigations (Beldade et al. 2011; Callahan et al. 2005; Lafuente and Beldade 2019; Scheiner 1993; Via et al. 1995).
In particular, body-size plasticity has been studied well, aiming to understand why ectotherms grow larger at lower temperatures, a process called the temperature-size rule (Angilletta and Dunham 2003; Atkinson 1994; Ghosh et al. 2013; Van Voorhies 1996). Atkinson (1994) gathered results on the temperature-size rules phenotype in ectotherms from an extensive number of studies and showed that 83% of the studies described that colder temperature resulted in significantly bigger body size. The same pattern of increased size at lower temperature was also observed in many insects and arthropods for body size and egg size (Azevedo et al. 1998, 2002; Czarnoleski et al. 2017; Ellers and Driessen 2011; Fischer et al. 2006; Klok and Harrison 2013; Steigenga et al. 2005). Although allelic variants and genes have been found to play an important role in body-size plasticity (Bochdanovits et al. 2003; Ghosh et al. 2013; Lafuente et al. 2018; Li et al. 2006), the genetic architecture underlying this phenomenon is not fully uncovered yet. This is because these traits plasticity are orchestrated by a complex interaction between genetics and environmental factors.
Nematodes are not exceptional to this phenomenon. For instance, the nematode Caenorhabditis elegans showed a 33% larger body size when grown at 10 °C compared to nematodes grown at 25 °C (Van Voorhies 1996) and other temperatures (i.e., 24 °C) (Gutteling et al. 2007b; Kammenga et al. 2007). Part of this phenotypic variation in lower-temperature-dependent body size was caused by natural genetic variation in the calpain-like protease tra-3 (Kammenga et al. 2007).
Overall, C. elegans is an attractive organism for studying the genetics of plasticity to temperature. Its small genome, rapid life cycle (3.5 days at 20 °C), genetic tractability, and a wealth of available experimental data have made this nematode a powerful platform to study the genetics underlying complex traits (Gaertner and Phillips 2010; Snoek et al. 2020). Besides, C. elegans can be maintained completely homozygous, produce many offspring (200–300 offspring per self-fertilizing hermaphrodite), and can be outcrossed with rarely occurring males (Petersen et al. 2015; Sterken et al. 2015; Gaertner and Phillips 2010). Furthermore, there are many temperature-related trait differences between two widely used divergent strains: N2 and CB4856. More specifically, studies reported that CB4856 and N2 differed in their response to temperatures in several life-history traits such as time to maturity, fertility, egg size, body size, lifespan, and also in gene expression regulation (Gutteling et al. 2007a, b; Jovic et al. 2017; Kammenga et al. 2007; Li et al. 2006; Rodriguez et al. 2012; Viñuela et al. 2011). Despite these findings, we still do not have a full overview of the loci that affect plasticity at a wider range of different temperatures.
To further elucidate the genetic architecture of temperature affected body-size plasticity in C. elegans, we selected 40 recombinant inbred lines (RILs) derived from N2 and CB4856 parents (Li et al. 2006) to study the plasticity and genetic regulation of body-size traits (body-size and some internal organs size) under four temperatures and at two developmental stages. First, we sought to investigate the effect of temperature and developmental stages on the reaction norms of the body-size traits, correlation between body-size traits within and between temperature-developmental stages, as well as investigating genetic parameters (heritability and transgressive segregation) of body-size traits and body-size plasticity. Subsequently, we investigated the genomic regions underlying these body-size traits across temperature-developmental stage combinations and plasticity traits under three temperature ranges. We found 18 quantitative trait loci (QTL) of body-size traits at a certain temperature and developmental stages and five plasticity QTL. Many of the QTL for different traits colocalized at the same position within temperatures suggesting a pleiotropic effect or close linkage. Furthermore, some of the plasticity QTL also colocalized with single temperature body-size QTL, suggesting a possibility of co-regulatory loci underlying plasticity traits and traits themselves. Moreover, the colocalizing QTL across temperatures indicates a possible temperature-sensitive regulatory mechanism.
Materials and methods
Mapping population
The mapping population used in this study consisted of 40 RILs from a 200 RIL population derived from the crossing of N2 and CB4856. These RILs were chosen because they were the smallest set of lines while capturing most of the genetic diversity within the larger set. The RILs were generated by (Li et al. 2006) and most were genotyped by sequencing, with a genetic map consisting of 729 informative (indicating a cross-over) single-nucleotide polymorphism markers (Thompson et al. 2015). The strain names and genotypes can be found in Supplementary Fig. S1.
We confirmed that long-range linkage, between markers on different chromosomes, was not present in the population, by studying the pairwise correlation of the genetic markers in the used population (Supplementary Fig. S2).
Cultivation and experimental procedures
C. elegans nematodes were reared following standard culturing practices (Brenner 1974). RILs were kept at 20 °C before experiments and 3 days before starting an experiment a starved population was transferred to a fresh NGM plate. An experiment was started by bleaching the egg-laying population, following standard protocols (Brenner 1974). After bleaching, nematodes were placed on a fresh NGM plate seeded with Escherichia coli OP50. From that point onward, RILs were grown at four different temperatures: 16, 20, 24, or 26 °C. At two timepoints of developmental stages (L4 and adult) per temperature, microscope pictures (Leica DM IRB, AxioVision) were taken of three nematodes per line per temperature that were mounted on agar pads. We decided on the amount of replication based on previous research, where it was found that the within-strain variance was not very high (Gutteling et al. 2007a, b). The timepoints were chosen such that L4 and young adult nematodes were photographed; this was confirmed by mid-L4 vulva shape and adult vulva shape and germline. The exact times are indicated in the sample data file (Supplementary Table S1).
Trait measurements and calculations
The number of RILs subjected to treatments per developmental stage was 40, except for treatment of temperature 24 °C at the L4 stage, where 39 RILs were used. Per life-stage and temperature, we took measurements of three replicate individuals per RIL, and six replicate individuals of the parental lines N2 and CB4856 (from two independent populations). This resulted in 1056 pictures.
To quantify traits, the pictures were loaded into ImageJ (version 1.51f) and traits were manually measured. In total, nine body-size traits were measured: (i) body length, (ii) width at vulva, (iii) length of the pharynx, (iv) width of the pharynx, (v) length of the isthmus, (vi) length of the buccal cavity, (vii) length of the procorpus, (viii) surface postbulb, and (ix) surface nematode. For surface postbulb and surface nematode, the perimeter of the trait was measured and the total surface area was calculated in ImageJ. To convert the measurement data from pixels to millimeters (mm), a figure of scale (in mm) was loaded to ImageJ. Subsequently, the resolution of C. elegans picture and the scale picture were equalized. Next, in ImageJ we determined how many pixels were represented by 0.1 mm. This step was repeated ten times and the average value was taken as a standard conversion scale from pixels to mm. We also calculated body volume (assuming the nematodes body resembles a tube) as follows:
and the length/width ratio (L/W ratio) as the ratio of body length/width at the vulva. For none of the traits we have a complete dataset due to difficulties in obtaining accurate measurements, the number of missing values for each trait are as follows: body length = 67; width at vulva = 102; length pharynx = 76; length isthmus = 220; surface postbulb = 219; surface nematode = 232; length buccal cavity = 193; length procorpus = 242; body volume = 135; width pharynx = 65; length/width ratio = 135. All raw data can be found in Supplementary Table S2.
Analytical software used
Phenotypic data were analyzed in “R” version 3.5.2 × 64 using custom-written scripts (R Core Team 2021). The script is accessible via Gitlab: https://git.wur.nl/published_papers/maulana_2021_4temp. R package used for organizing data was the tidyverse (Wickham et al. 2019), while all plots were made using ggplot2 package (Wickham 2011), except for heatmaps in Supplementary Fig. S3 that were made using the “heatmap ()” function provided in R. The data were deposited to WormQTL2 where it can be explored interactively (www.bioinformatics.nl/WormQTL2) (Snoek et al. 2020).
Correlation analysis
The correlation between the traits in all treatment combinations was determined by the Pearson correlation index and plotted in a correlation plot. To diminish the effects of very high and low values of single observations, we normalized the data as follows:
where x is an individual observation of the traits in temperature i (16, 20, 24, 26 °C) and developmental stage j (L4, adult) while µ is the mean value of all traits.
Transgressive segregation
To determine transgressive segregation of the traits among RILs panel, we performed multiple t-tests comparing all RIL panels to both parents for all traits per temperature and developmental stages. Transgression was defined when the traits of individual RIL is significantly different than both parents (p.adjust with false-discovery rate (FDR) < 0.05; equal variance not assumed).
Heritability estimation
Broad-sense and narrow-sense heritability of the phenotypic traits over RIL lines was calculated using the restricted maximum likelihood (REML) model to explain the variation of the traits across the RIL lines (Kang et al. 2008; Rockman et al. 2010). The broad-sense heritability was calculated according to the following equation:
where H2 is the broad-sense heritability, Vg is the genotypic variation explained by the RILs, and Ve is residual variation. The Vg and Ve were estimated by the lme4 model xnorm ~ 1 + (1|strain) (Bates et al. 2015). The input data were normalized using log2 phenotype value to better fit the normality assumption.
Narrow-sense heritability is defined as the total variation in the population that is captured by additive effects. We calculated these using the heritability package in R, which estimates narrow-sense heritability based on a kinship matrix (Kruijer et al. 2014). The kinship matrix was calculated using the kinship function from the Emma package in R (Carta et al. 2011).
The significances of broad and narrow-sense heritability were determined by permutation analysis where the traits values were randomly assigned to the RILs. Over these permutated values, the variation captured by genotype and residuals were then calculated. This permutation was done 1000 times for each trait. The results obtained were used as the by-chance-distribution and an FDR = 0.05 threshold was taken as the 50th highest value.
QTL mapping
QTL mapping was performed using a custom script in R using fitted single marker model as follows:
where µ is the average of all strains replicates in terms of their body-size traits i, of RIL j (N = 40) on marker location x (x = 1, 2, 3, …., 729).
Detection of QTL was done by calculating a –log10(p) score for each marker and each trait. To increase the detection power, all the values were log2 normalized. To estimate the empirical significance of –log10(p), the traits were randomly permutated values over the RILs 1000 times. The calculation resulted in a significance threshold with FDR = 0.05 at a –log10(p) of 3.4 for QTL detection.
Trait plasticity calculation
We divided plasticity ranges into three adjacent temperature groups: 16–20 °C, 20–24 °C, and 24–26 °C. Trait plasticity was defined as a ratio between the trait mean value per nematode strain at 16–20 °C, 20–24 °C, and 24–26 °C.
Heritability estimation of trait plasticity
Broad-sense and narrow-sense heritability of trait plasticity was calculated using the same model as in phenotypic traits heritability above. The broad-sense heritability was calculated according to the following equation:
where Hp2 is the broad-sense heritability of trait plasticity, Vg is the genotypic variation explained by the RILs, and Ve is residual variation. The Vg and Ve were estimated by the lme4 model xnorm ~ 1 + (1|strain) (Bates et al. 2015).
The narrow-sense heritability was estimated based on the kinship matrix calculated using the kinship function from the Emma package in R (Carta et al. 2011). The significances of broad and narrow-sense heritability were determined by permutation analysis as in phenotypic heritability estimation. The calculation was done for all temperature ranges (16–20 °C, 20–24 °C, and 24–26 °C) in the adult and L4 stage.
QTL mapping for trait plasticity
Plasticity QTL mapping was performed using a fitted single marker model as follows:
where µ is the average of all strains replicates in terms of their body-size traits i, of RIL j (N = 40) on marker location x (x = 1, 2, 3, …., 729).
Plasticity QTL detection was done by calculating a –log10(p) score for each marker and each trait. The calculation was done per temperature ranges. Empirical significance of –log10(p) was estimated by randomly permutated value over the RILs 1000 times. The FDR = 0.05 at a –log10(p) of 3.0 was found as the significant threshold for plasticity QTL detection.
Statistical power calculation
To determine the statistical power of our QTL and plasticity QTL dataset at the set threshold, we performed power analysis using the genetic map of the strains (n = 40) used per condition as in (Sterken et al. 2017). We simulated ten QTL per marker location that explained 20–80% of the variance, with increments of 5% (20, 25, 30,…, 80%). Random variation was introduced based on a normal distribution with σ = 1 and µ = 0. Peaks were simulated according to effect-size, for example, a peak corresponding to 20% explained variation was simulated in this random variation. Based on the simulation, we analyzed the number of correctly detected QTL, the number of false positives as well as undetected QTL. In addition, the precision of effect-size prediction and the QTL location were determined. The threshold used was based on 1000× permutation analysis –log10(p) > 3.4 for individual QTL and –log10(p) > 3.0 for plasticity QTL. The results of this calculation are presented in Supplementary Table S5.
Results
C. elegans body-size traits vary across temperatures and developmental stages
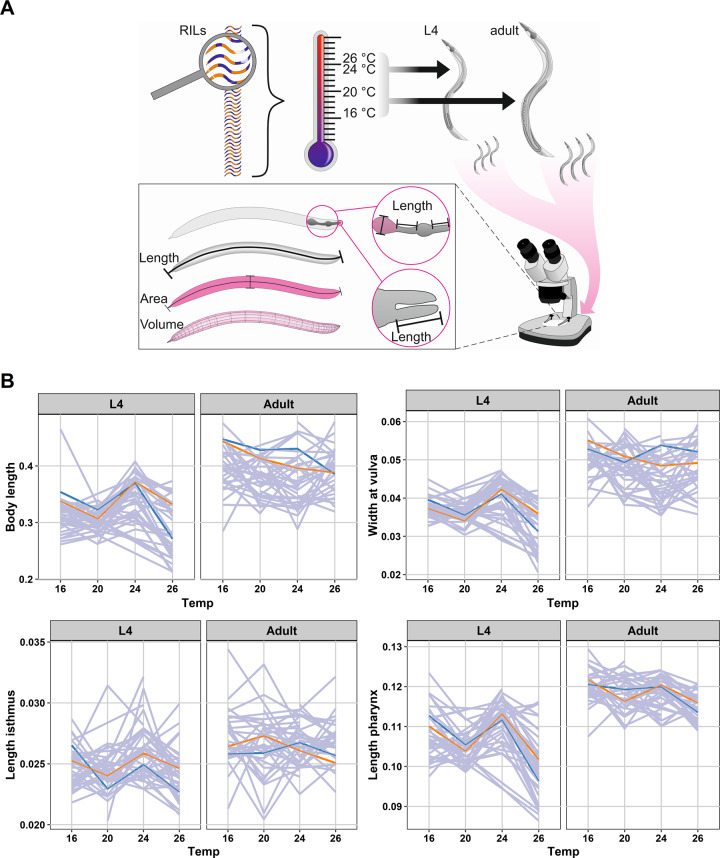
To investigate the impact of different genetic background, ambient temperature condition, and developmental stages on the body-size traits, we used a panel of 40 RILs derived from a cross between Bristol strain (N2) and Hawaiian strain (CB4856) (Supplementary Figs. S1 and S2) (Li et al. 2006). Each individual RIL was grown under four different temperature regimes (16, 20, 24, and 26 °C). Once reaching the L4 and adult stage, we took pictures of each RIL with three individual replicates per strain and determined the body-size parameters using ImageJ (Fig. 1A; see Materials and methods).
Fig. 1. Body-size variation on four temperature regimes in C. elegans RILs.
A Flowchart of experimental overview. A set of 40 RILs was grown at temperatures 16, 20, 24, and 26 °C. Separately, at the L4 and adult stages, individual RIL with three replicates per RIL was photographed under the microscope. Subsequently, the body-size traits of the RILs were determined using ImageJ. B Reaction norms of four C. elegans body-size traits plasticity across different temperature and developmental stages. The x-axis represents temperatures used while y-axis represents the mean value of the individual strains in their respective traits (in mm). Both parents are depicted in blue (CB4856) and orange (N2), while the RILs are gray.
The 11 body-size traits showed variation across RILs when measured in different temperatures and developmental stages (Fig. 1B and Supplementary Fig. S3), suggesting that these traits are indeed plastic. In general, we observed a similar dynamic pattern of traits plasticity across RILs in the L4 stage including N2 and CB4856. The trait values dropped from 16 to 20 °C, then increased from 20 to 24 °C, and dropped again from 24 to 26 °C. On the other hand, the trait values in adult stages show no similar pattern across the RILs, suggesting that the traits plasticity in the adult stage are more sensitive to genetic background, whereas in the larvae the environment seems to play a larger role (Fig. 1B and Supplementary Fig. S3). For several major body-size traits such as body length, body volume, width at vulva, and surface area of the nematodes of adult worms, we found that CB4856 did not completely follow the temperature-size rules (a decreasing curve from 16 to 20 °C, followed by an increase from 20 to 24 °C, and decrease from 24 to 26 °C) whereas Bristol N2 consistently grew bigger at lower temperatures. These results complement previous findings on the plasticity of body-size over two temperature ranges (Gutteling et al. 2007b; Kammenga et al. 2007). To get insight into the relations between the traits measured, we performed a correlation analysis for all pairs of traits at the two developmental stages. We found that the level of between trait-correlation differed between L4 and adult stages, where the temperature seems to be the main driving factor (Supplementary Fig. S4). Both in the L4 and adult stage, the body-size traits displayed a strong positive correlation within the same temperature, and a strong negative correlation between different temperatures, suggesting that the variation in the body-size traits were temperature specific. Interestingly, both in the L4 and adult stages, the body-size traits of worm grown in 16 and 26 °C were separated into several small clusters, while the traits from 20 to 24 °C treatments formed a single positively correlated cluster. These results indicated that there were more similar patterns of variation over RILs in temperature 20 and 24 °C. Strongly correlated body-size traits imply that the same QTL could be detected for these traits due to similar patterns of variation in the RILs, temperatures, and developmental stages.
To explore the source of variation of the body-size traits in the RILs population, we used principal component analysis (PCA) (Supplementary Fig. S5). The PCAs describe the variation of the traits based on temperatures and genetic background per developmental stage. At the L4 stage, the first principal component captured 45.5% of the variation where the 16 °C temperature animals were most distal from the other temperatures, while the second principal component captured 24% of the variation where the 24 and 26 °C temperatures were most distal. We found that at the L4 stage, the RILs were more similar in lower temperature (16 °C) while at 20 °C, they were distributed across the PC plot. Subsequently, the value of body-size traits of the nematodes at 24 °C was similar to the values at 26 °C, but divided into two clusters (Supplementary Fig. S5). On the other hand, the individual RILs did not show any clear clusters at the adult stage, indicating there was high variation between the RILs as a result of interaction between the environment and the genetic background. This result combined with the correlation analysis shows that there was a substantial variation in the RILs, suggesting that it was possible to detect QTL controlling the traits.
Transgressive segregation and heritability indicate a complex genetic architecture underlying body-size traits
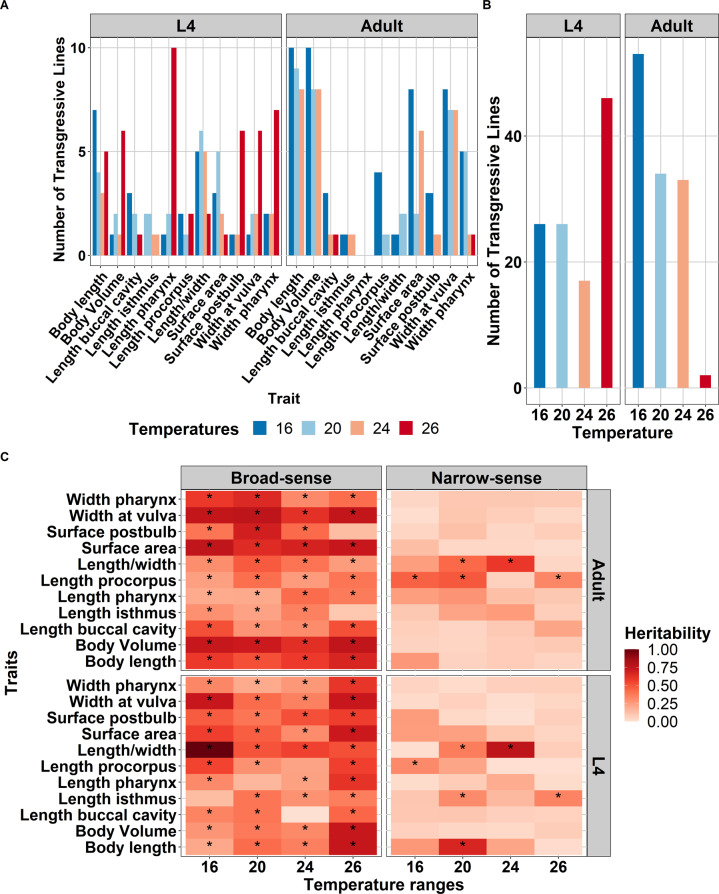
Upon inspecting the distribution of trait variation in the RILs compared to N2 and CB4856, we observed high levels of variation exceeding those of the parental strains (Supplementary Fig. S6). This suggests transgressive segregation within the RIL population. Hence, we tested the trait values of each RIL versus the parents. We found transgression for almost all traits per temperature-developmental stage combinations (t-test, p.adjust FDR < 0.05) (Supplementary Table S3). Our findings show that the number of two-sided transgressive RILs depended on the combination of temperature and developmental stage (Fig. 2A, B and Supplementary Table S3). In the L4 stage, the number of transgressive RILs was constant under 16 and 20 °C, slightly dropped under 24 °C, and then increased at 26 °C. Conversely, in the adult stage, the number of transgressive strains decreased as the temperature increased. Moreover, it shows that the parental lines have both positive and negative alleles that interact with the environment leading to a more robust/stable phenotype over a broader temperature range. Using ANOVA, we found that developmental stage was indeed the factor driving transgression (p = 0.0275; R2 = 0.073; Table 1), whereas temperature alone showed no relation to the transgression (p = 0.786; R2 = 0.015). These findings indicate the environment- and age-specific effects on the regulation of body-size traits.
Fig. 2. Transgression and heritability for body-size traits on four temperature regimes.
A The number of transgressive lines of 11 body-size traits in C. elegans across temperatures and developmental stages. The traits are on the x-axis while y-axis represents the number of transgressive lines based on multiple t-test of traits in each individual line against both parents (p.adjust FDR < 0.05). Colors represent the temperatures treatment, corresponding to the legend on the bottom side. B The number of transgressive traits found in the RIL population per treatments combination (temperature-developmental stage). The temperature is on the x-axis while y-axis represents how many transgressions were found within those temperatures. Developmental stages are depicted on the above side of the graph. C Broad-sense and narrow-sense heritability of body-size traits across temperature and developmental stages. On the x-axis are temperature and on the y-axis are the traits measured. The color gradient represents the heritability values as depicted on the legend on the right side of the plot. Asterisk (*) inside the box indicate significant heritability values (FDR0.05 based on 1000 permutations). Developmental stages are depicted on the right side of the plot whereas the types of heritability are on the top.
Table 1.
Results of ANOVA for the number of transgressive lines over temperatures and developmental stages.
| Source | Df | Sum Sq | Mean Sq | F value | Pr(>F) | R2 |
|---|---|---|---|---|---|---|
| Temperature | 3 | 7.7 | 2.55 | 0.35 | 0.787 | 0.015 |
| Developmental stage | 1 | 37.0 | 37.01 | 5.13 | 0.028 | 0.073 |
| Temperature × Developmental stage | 3 | 52.3 | 17.43 | 2.41 | 0.076 | 0.103 |
| Residuals | 56 | 404.4 | 7.22 |
ANOVA analysis of variance, Df degree of freedom.
Next, to determine the proportion of variance in body-size traits that were caused by genetic factors, we calculated the broad-sense heritability (H2) of each trait. We found significant heritability (REML, FDR = 0.05) for 81 out of 88 traits in developmental stage-temperature combinations. The significant heritability ranged from 0.202 (width pharynx at 20 °C in L4) to 0.99 (length/width ratio at 16 °C in L4) (Fig. 2C and Supplementary Table S4). Hence, for a large fraction of traits, we could detect a high contribution of genetic factors. In addition to H2, we calculated the narrow-sense heritability (h2) to identify how much of the variation could be explained by additive allelic effects. This analysis suggested that there were 11 traits with significant additive effect (REML, FDR < 0.05; Supplementary Table S4). For nearly all body-size traits we detected H2 well beyond h2, indicating a role for epistasis in the genetic architecture of the traits.
To understand the contribution of temperature and developmental stage on heritability of all traits measured, we conducted an ANOVA (Table 2). The results suggested a trend that the main factor driving H2 was the interaction between temperature and developmental stages (R2 = 0.084, p = 0.066). On the other hand, temperatures and developmental stages by themselves showed little relation to the variation of H2 (R2 = 0.008 and R2 = 0.005, respectively). In the adult stage, we observed there were four traits (width at vulva, body length, body volume, and surface area of nematodes) which H2 are relatively robust across all temperatures while in L4 they were found to be more variable across temperature. These four traits were affecting each other and were observed to be positively correlated (Supplementary Fig. S3). Taken together, overall body-size traits show significant H2, indicating a substantial effect of the genetic background on the variation in these traits in this population. Moreover, the correlation between some traits indicates a shared genetic architecture between the traits. These results indicate a higher chance of detecting QTL on the traits measured.
Table 2.
Results of ANOVA for H2 of all traits over temperatures and developmental stages.
| Source | Df | Sum Sq | Mean Sq | F value | Pr(>F) | R2 |
|---|---|---|---|---|---|---|
| Temperatures | 3 | 0.292 | 0.097 | 2.35 | 0.078 | 0.008 |
| Developmental stages | 1 | 0.019 | 0.019 | 0.472 | 0.494 | 0.005 |
| Temperatures × Developmental stage | 3 | 0.308 | 0.103 | 2.487 | 0.066 | 0.084 |
| Residuals | 80 | 3.308 | 0.041 |
The input data used were the heritability measurements of all traits.
ANOVA analysis of variance, Df degree of freedom.
QTL underlying body-size traits in C. elegans are influenced by temperature and developmental stages
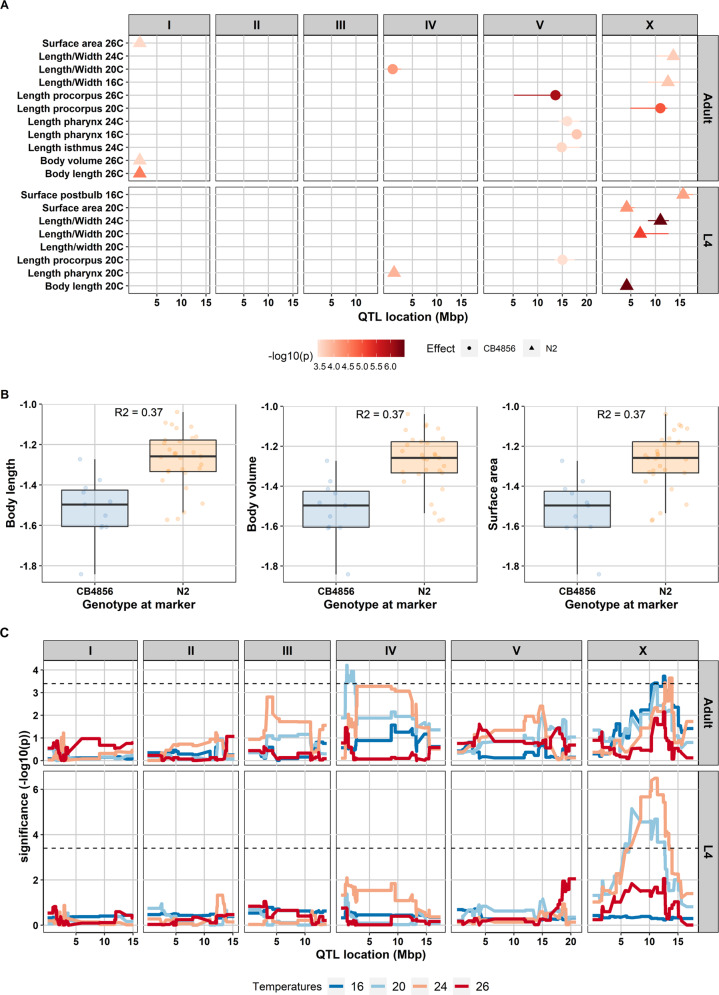
To identify underlying loci controlling the variation of body-size traits, we performed QTL mapping for all the body-size traits measured in the 40 RILs. Analysis of statistical power showed that our population can detect 80% of true QTL explaining 60% of the variance (Supplementary Table S5). Using log-normalized mean values per RIL as input, we found 18 significant QTL (–log10(p) = 3.4, FDR = 0.05) with –log10(p) scores ranging up to 6.5 in each temperature and developmental stages (Fig. 3 and Supplementary Table S6). We found QTL explaining 28–53% of the variance among the RILs (Supplementary Table S6). We found seven QTL in the L4 stage, namely, surface area, length pharynx, body length, length procorpus (detected at 20 °C), length/width ratio (detected at 20 and 24 °C), and surface postbulb (detected at 16 °C). For the adult stage, 11 QTL were detected for the body-size traits. Here, we found QTL evenly distributed over the temperatures: two at 16 °C, two at 20 °C, three at 24 °C, and four at 26 °C (Fig. 3A). Of the 18 significant QTL, 8 were located on chromosome X, 5 QTL on chromosome V, 3 on chromosome I, and 2 on chromosome IV.
Fig. 3. Quantitative trait locus mapping of body-size traits in four temperature regimes.
A QTLs found for body-size traits in the 40 RILs. The x-axis represents the position of the QTL in mega base pairs (Mbp) for each chromosome and y-axis displays the corresponding significant QTL from a single marker model. In total, 18 QTL were found with –log10(p) score ranging from 3.44 to 6.49 (–log10(p) threshold 3.4, FDR = 0.05). Shapes represent genotype effect: dots = CB4856; triangles = N2. Horizontal lines in the QTL spots represent confidence interval. B Allelic effects of QTL for body length, body volume, and surface area of the nematode in adult stage at 26 °C at the peak marker location. RILs that have N2 marker at this locus relatively have a bigger body size compared to those that have CB4856 marker. The genetic variation on chromosome I can explain 30–37% of the variation in body length, body volume, and surface nematode at that condition. C QTL profile of length/width ratio. The QTL analysis was performed across four temperatures (16, 20, 24, 26 °C) and two developmental stages (L4 and adult) using a single marker model. X-axis displays genomic position in the chromosome corresponding to the box above the line while y-axis represents the –log10(p) score. Box in the right graph shows the developmental stages. Blue line represents QTL at 16 °C, light blue line at 20 °C, orange line at 24 °C, and red line at 26 °C. Black-dash line represents –log10(p) threshold (FDR = 0.05).
We observed QTL-hotspots for various traits. For example, the chromosome I QTL (surface area, body volume, and body length) were positively correlated traits, mapped in the same developmental stage and temperature combination. Hence, this could point to a body-size QTL, where the N2 genotype was associated with a larger body size compared to the CB4856 genotype (Fig. 3B). Interestingly, all QTL on chromosome V were associated with the size of the feeding apparatus, were found over various temperatures, and were all associated with increased size in CB4856 (Fig. 3A and Supplementary Fig. S7). In contrast, traits related to the overall body size (e.g., volume) were almost exclusively associated with an increased size due to the N2 allele (Fig. 3B).
In line with the indications of the correlation- and heritability analyses, we found evidence for environment (temperatures), age (developmental stage) and genotype interactions. For example, for length/width ratio (Fig. 3C) in the adult stage, significant QTL in chromosome X were detected for the worms grown at 16 and 24 °C, one QTL on chromosome IV for worms grown at 20 °C, and no significant QTL detected at 26 °C. When we mapped the trait in adult worms grown at 16 °C, we found a significant QTL on chromosome X that we did not find in the L4 stage at 16 °C. The same result was found for QTL at temperature 20 °C at the adult stage on chromosome IV that was not present in the L4 stage. Similar patterns of (dis-) appearance were observed for many traits (Supplementary Fig. S7). Hence, traits may be regulated by different sets of genes (loci) dependent on temperature-environment and developmental stage. This indicates that there is a considerable effect of genotype-environment interactions.
The RIL population revealed plasticity QTL for several body-size traits
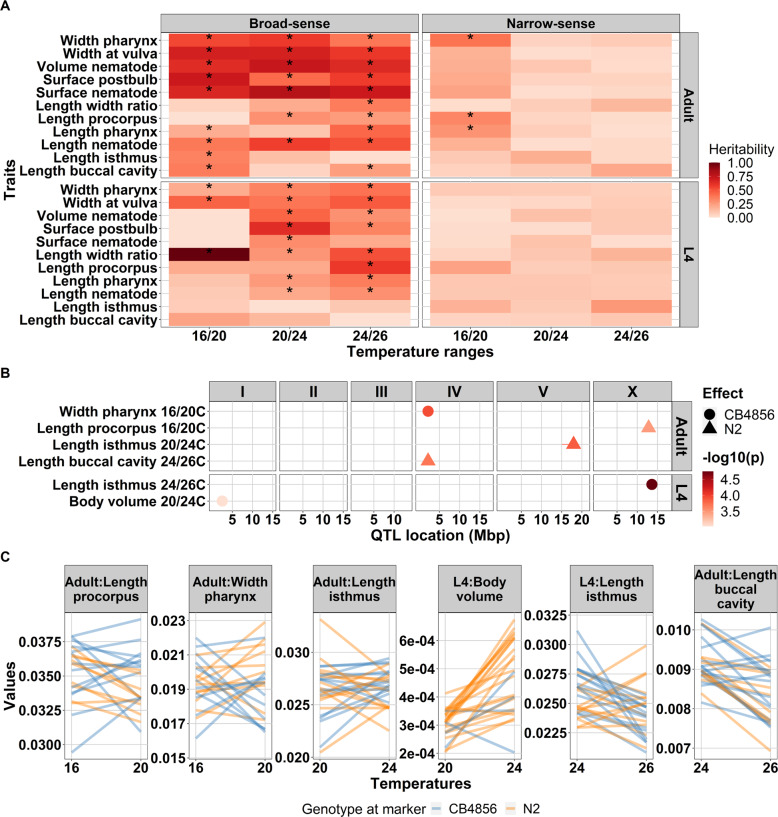
Phenotypic plasticity is the change of the expressed phenotype in different environments. To determine the amount of variation in body-size plasticity was due to genetic factors, we calculated the H2 of each set of neighboring experimental temperatures: we defined plasticity as the ratio of the traits in 16–20 °C, 20–24 °C, and 24–26 °C. We found significant H2 (REML, FDR = 0.05) for trait plasticity for 45 out of 66 traits in developmental stage-temperature ranges combinations. In contrast, for h2 of traits plasticity, there were only three traits (length pharynx, length procorpus, and width pharynx, and adult stage on temperature ranges of 16–20 °C) with significant additive effect (REML, FDR < 0.05; Supplementary Table S7). The significant h2 values ranged from 0.19 (length pharynx at adult stage on 20–24 °C range) to 1.00 (length/width ratio on 16–20 °C in L4 stage) (Fig. 4A and Supplementary Table S7). Consistent with the patterns observed across temperatures in the L4 versus the adult stages (Fig. 1B), there were fewer significant broad- and narrow-sense heritabilities observed in the L4 stage compared to the adult stage, indicating higher environmental variation of traits plasticity in L4. In summary, we observed a strong effect of developmental stages on the heritability of trait plasticity which was also strongly dependent on the body-size trait under study (Table 3).
Fig. 4. Quantitative genetic variation in trait plasticity.
A Broad-sense and narrow-sense heritability of body-size traits plasticity across temperature range and developmental stages. On the x-axis temperature plasticity and y-axis are the traits measured. Color gradient represents the heritability value as depicted on the legend on the right side of the plot. Asterisk (*) inside the box represent a significant heritability value (FDR0.05 based on 1000× permutation). Developmental stages are depicted on the right side of the plot whereas the type of heritability is on the top. B Plasticity QTLs found for body-size traits in the 40 RILs. The x-axis represents the position of the QTL in million base pairs (Mbp) for each chromosome and y-axis displays the corresponding significant plasticity QTL based on a single marker model. In total, five plasticity QTL were found with –log10(p) score ranging from 3.03 to 4.75 (–log10(p) threshold 3.0. Horizontal lines in the QTL spots represent confidence interval. C Phenotypic values (in mm) of the corresponding plasticity QTL. X-axis represents the temperature regime where the plasticity QTL identified. Y-axis represents the mean phenotypic values of the traits. Orange lines were RILs with N2 genotype at the peak marker location, whereas blue lines represent RILs with CB4856 genotype at the marker location. The traits are depicted on the top of each plot.
Table 3.
Results of ANOVA for H2 of all trait plasticity over temperatures and developmental stages.
| Source | Df | Sum Sq | Mean Sq | F value | Pr(>F) | R2 |
|---|---|---|---|---|---|---|
| Temperatures ranges | 2 | 0.044 | 0.022 | 0.407 | 0.6677 | 0.012 |
| Developmental stages | 1 | 0.340 | 0.34 | 6.234 | 0.0153 | 0.092 |
| Temperatures ranges × Developmental stage | 2 | 0.030 | 0.015 | 0.275 | 0.7605 | 0.008 |
| Residuals | 60 | 3.272 | 0.0545 |
The input data used were the heritability measurements of all traits.
ANOVA analysis of variance, Df degree of freedom.
In contrast to H2 values per temperature-trait combination where the main factor driving the heritability was temperature, in H2 of plasticity, developmental stage (R2 = 0.092, p = 0.0153) was the main explanatory factor. On the other hand, temperature ranges showed little relation to the variation of plasticity H2 (R2 = 0.012, p = 0.667). Taken together, overall body-size traits plasticity showed significant H2, indicating a substantial effect of the genetic background on the variation of these traits in this population.
The previous results suggested that QTL affecting body-size traits can be located on different chromosomes when measured in a different environment, indicating an environment-QTL interaction. To further understand the mechanism of trait plasticity, we mapped QTL for trait plasticity. Statistical power analysis indicated that we can detect 80% of QTL explaining 60% of variation (Supplementary Table S5). For all three conditions, we found six significant plasticity QTL (Fig. 4B and Supplementary Fig. S9). Two plasticity QTL were found in the temperature range from 16 to 20 °C harboring a locus associated with width pharynx at the adult stage, and length of the procorpus at the adult stage. Two plasticity QTL in a temperature range of 20–24 °C were associated with length isthmus at the adult stage and body volume at the L4 stage. Two plasticity QTL in temperature ranges of 24–26 °C related to length isthmus at L4 stage and length buccal cavity at adult stage (Fig. 4, BC and Supplementary Table S8). Hence, we found less QTL than for the individual temperatures. However, this was to be expected since the narrow-sense heritability of plasticity was lower and resulted in fewer significant values.
We compared the plasticity QTL to the QTL mapped for the individual temperatures. Of the six plasticity QTL, three of them were precisely colocalized with QTL for body-size traits within individual temperatures (Figs. 3A and 4B): (1) plasticity QTL of length procorpus in the temperature range from 16 to 20 °C colocalized with QTL for length/width ratio of adult nematode at 16 °C; (2) plasticity QTL for length isthmus of L4 worms in temperature ranges of 24–26 °C colocalized with length/width ratio of adult worms in 24 °C; (3) plasticity QTL for length isthmus of adult worms in the temperature range of 20–24 °C colocalized with QTL for length pharynx at the adult stage at 16 °C. These results indicate that although plasticity can be reflected in individual temperatures, contrasting trait values over varying conditions reveals new insights into the underlying loci.
We continued by investigating the direction of the plasticity QTL. In three of the QTL the N2 genotype had a negative effect on the trait value, namely for length procorpus (16–20 °C at the adult stage), length isthmus (20–24 °C at the adult stage), and length buccal cavity (24–26 °C at the adult stage). In other words, the RILs harboring N2 genotype at the peak marker location have decreased phenotype, while CB4856 genotype has an increased phenotype (Fig. 4C). In contrast, for plasticity QTL of width pharynx (16–20 °C at the adult stage), body volume (20–24 °C at L4 stage), and length isthmus (24–26 °C at L4 stage), the RILs with an N2 locus display an increase of the trait value. The slope of reaction norm of the trait plasticity indicates allele(s) that affect the trait variation both in environment 1 and environment 2 linked to genetic variation in plasticity (Lafuente and Beldade 2019). Taken together, the heritability analyses indicated that in general, temperature-related plasticity was regulated by complex genetic effects over the course of the gradient. The lack of mapped (large-effect) QTL might indicate that underlying loci act in a gradual rather than a sudden way.
Discussion
Body-size traits reaction norm reveals a genotype × environment interaction
In most ectotherms, temperature is an important factor driving body size and is related to life-history traits (Angilletta and Dunham 2003; Ellers and Driessen 2011; Ghosh et al. 2013; Peng et al. 2007). This was also found for C. elegans (Gutteling et al. 2007a, b; Kammenga et al. 2007). In this study, we used a C. elegans RIL population to study the underlying genetic regions that regulate the body-size traits, both main body traits (i.e., length, width at vulva, volume, length/width ratio, and surface area) as well as internal organs and feeding apparatus (i.e., length isthmus, length procorpus, length pharynx, width pharynx, and surface postbulb), and the plasticity of such traits in different temperatures at two developmental stages.
We observed that the reaction norm of the body-size traits over temperatures were varied depending on the trait and individual genotypes and showed a clear genotype-environment interaction (G×E) (Beldade et al. 2011; Lafuente and Beldade 2019; Lafuente et al. 2018; Saltz et al. 2018). The fact that some RILs do not follow the temperature-size rule, especially for major body-size traits, might be the result of CB4856 genotype in the RILs, as this strain is known to deviate from this rule (Gutteling et al. 2007a, b; Kammenga et al. 2007). Interestingly, we also found that the shape of reaction norm was affected by the developmental stage of the animal. For most of the traits measured, adult worms displayed non-parallel reaction norm across temperature, as opposed to L4 stage worms that showed relatively similar non-linear parallel reaction among genotypes in most of the traits (Supplementary Fig. S4). This indicates that variation in phenotypic plasticity was not common in juvenile animals suggesting the absence of (or less significant) GxE for most genotypes as described in (Saltz et al. 2018). These differences of L4 and adult stage reaction norm could stem from differences in interaction between gene-expressions and environment (temperature) in L4 and adult (Lafuente and Beldade 2019; Li et al. 2006; Snoek et al. 2017; Viñuela et al. 2010).
Within C. elegans it was previously known that the reaction norm of body length and body volume was defied in the CB4856 strain due to a polymorphism in tra-3 (Gutteling et al. 2007b; Kammenga et al. 2007). This was measured over a two-temperature gradient from 12 to 24 °C. We found that the body size of CB4856 was dynamic (decrease and increase) over the course of temperatures. Meanwhile, for N2, we found that the negative linear norm was most apparent from 16 to 20, and at higher temperatures the overall body-size (e.g., length, width at vulva, surface area, and volume) were robust. It suggests that the body-size trait of N2 follows the “threshold” reaction norm model, instead of the linear function as described in (Lafuente and Beldade 2019). This discrepancy could only be revealed by using four temperatures (16, 20, 24, and 26 °C) as in this study.
Previous studies have suggested the effect of temperatures on genetic correlation of body size and several life-history traits in different species. Lafuente et al. (2018) found that body size (thorax and abdomen) of Drosophila melanogaster reared at 17 and 28 °C significantly correlates with chill coma recovery and survival of Metarhizium anisopliae fungi. Norry and Loeschcke (2002) found a positive correlation effect of lifespan with temperature and sex in D. melanogaster at 25 °C where the male flies lived longer. However, this effect was reversed at 14 °C. In C. elegans, an 18% of increase lifespan due to heat-shock was reported for CB4856 but not for N2, whereas the RILs showed a wide range of lifespan variation (Rodriguez et al. 2012). Our results are in agreement with other previous studies (reviewed in Sgro and Hoffmann 2004) that different environmental conditions result in a different correlation power, suggesting that evolutionary trajectories on trade-offs between traits, especially the traits that are controlled by specific loci, depend strongly on the environmental condition.
Genetic parameters and QTL analysis indicate a complex genetic regulation of body-size traits
In a population derived from two diverse parents, it is common to detect extreme phenotypes exceeding way beyond the parents (transgression) (Rieseberg et al. 1999). Transgression can represent the genetic complexity of a trait, for example, due to genetic interaction (epistasis) or it could mean that the trait is controlled by multiple loci with opposite effect combinations in the parental strains resulting in a similar phenotype. Transgression has been reported for C. elegans life-history traits such as egg size, number of eggs, body length (Andersen et al. 2015; Gutteling et al. 2007b; Kammenga et al. 2007), lifespan (Rodriguez et al. 2012), as well as metabolite levels (Gao et al. 2018), and gene expression (Li et al. 2006). We found transgressive segregation for almost all traits in temperature and developmental stage combinations, indicating a complementary action of multiple loci underlying these traits.
We then calculated the broad-sense heritability to investigate the proportion of variance explained by genetic factors in our RIL population. It should be noted that because of the necessity of using batches, H2 represents an upper-bound. Still, our estimation of broad-sense heritability of adult body length at 20 °C (0.51) was similar (H2 = 0.57) as reported by Andersen et al. (2014). In addition, the H2 of body volume (0.71) and width at vulva (0.76) in this study are also similar to (Snoek et al. 2019), which were 0.77 and 0.75, respectively. Heritability is a population trait characteristic and highly depends on the type of population used and the environment. Therefore, the fact that we found similar heritability with previous works indicates that the variation of these traits is quite stable between different mapping population. This could also mean that the relative effect of the micro-environment as well as the stochasticity is small. Furthermore, similar patterns of heritability that changed over temperatures (12 and 24 °C) were reported for body mass (volume), growth rate (change in body length), age at maturity, egg size, and egg numbers (Gutteling et al. 2007a, b).
By QTL mapping, we found 18 significant QTL for 88 temperature and developmental stage combinations regulating body-size traits. Here, we showed that QTL of some of the traits were colocalized in the same location in chromosome. For example, body length and surface area of the nematode in the L4 stage at 20 °C shared the same genomic region on the left arm of chromosome X. This is expected since these traits have a strong positive correlation. All the colocalized traits showed the same QTL effect where N2-derived loci were associated with an increase in size. It is possible that such colocalized QTL were the result of a single pleiotropic modifier affecting various aspects of the C. elegans physiology. On the other hand, this might be the result of unresolved separate QTL (Dupuis and Siegmund 1999; Gutteling et al. 2007b; Sterken et al. 2020).
As body length at 20 °C has been investigated across multiple studies, we used it to cross-reference our mapping. The same location (chromosome X: 4.9 Mb) was mapped in two other studies (Andersen et al. 2014, 2015). Furthermore, many QTL located in the left arm of chromosome X were associated with body length, indicating the alleles controlling these traits might the same or linked with alleles of body length. In another study, using a multi-parent RIL, it was found that loci located in the same position (chromosome X around 4.5–5 Mb) were associated with length/width ratio, which is also related to body length (Snoek et al. 2019). For the same trait, Snoek et al. (2014) found the QTL in different chromosomes (i.e., chr IV), meaning that our study has the power to reveal the previous undetected QTL.
Nagashima et al. (2017) summarized factors and its genetic basis involved in regulating body size in C. elegans including DBL-1, TGF-β signaling, DAF-2, rict-1, sma-5, wts-1, IGF-signaling, tra-3, npr-1, cat-2, dop-3, eat-2, pha-2, and pha-3. We manually checked the position of those genes in www.wormbase.org and found that none of the genes located under our QTL, except for npr-1 (wormbase 2021). Our QTL in chromosome X overlapped with the location of the Neuropeptide receptor 1 (npr-1) allele, which encodes the mammalian neuropeptide Y receptor homolog. This allele is a known pleiotropic regulator affecting traits such as lifetime fecundity, body size, and resistance to pathogens mediated by altered exposure to bacterial food (Andersen et al. 2014; Nakad et al. 2016; Reddy et al. 2009; Sterken et al. 2015). Moreover, seven out of eight QTL that colocalized in chromosome X have an increased size that is associated with N2 genotype, which supports our hypothesis that those seven traits could be npr-1 regulated.
Although not significant, we found potential QTL of body volume and width at vulva of adult nematode at 20 and 24 °C on the left arm of chromosome IV (Supplementary Fig. S7) that overlapped with QTL identified previously for body volume by (Gutteling et al. 2007b; Kammenga et al. 2007) at 24 °C using a larger population of RILs, also in chromosome X at 20 °C using multi-parental RIL (Snoek et al. 2019). These results indicate that these QTL represent robust and predictable genetic associations with temperature and size.
From 18 significant QTL, 9/18 (50%) were transgressive and 15/18 (83%) of the QTL had moderate to high heritability (>0.3). These findings indicate a highly complex genetic regulation of many body-size traits that could involve multiple interactions of different genetic variants. This was supported by the higher value of broad-sense heritability compared to narrow-sense heritability that suggests that the driving factors of most heritable traits were additive loci of opposing effects or genetic interactions.
Mapping of plasticity increments indicates small effect-size changes resulting in shifting loci
By mapping phenotypic plasticity over adjacent temperatures, we only found five plasticity QTL. We found two plasticity QTL over 16–20 °C that were related to width pharynx and length procorpus, both in the adult stage. In addition, we detected two plasticity QTL over 20–24 °C that was related to length isthmus in the adult stage and body volume at the L4 stage. Lastly, one plasticity QTL over 24–26 °C was associated with the length isthmus at the L4 stage. These results suggest that the QTL associated plasticity was environment specific, meaning that the candidate genes in the QTL region are differentially expressed depending on environmental conditions (Gutteling et al. 2007b).
We found little overlap between QTL for trait plasticity and QTL of traits in specific temperatures and developmental stages. Moreover, the plasticity QTL and traits QTL that colocalized were related to different traits. This low overlap of plasticity QTL and body-size trait QTL was also reported for D. melanogaster (Lafuente et al. 2018). Our results contribute to the long-standing debate on the genetic basis of plasticity (whether it is controlled via specific loci for trait plasticity or via the same loci that regulate trait in a certain environment) (Via et al. 1995). We showed that the genetic basis of trait plasticity, to some extent, can be the same as the genetic basis of traits in a certain environment, which support both ideas in congruence with previous papers, e.g., Têtard-jones et al. (2011). We also showed that one locus can be responsible for different traits as well as responsible for plasticity in different environments. These findings may also indicate an allelic sensitivity model underlying plasticity mechanism where loci display environmental-based allelic sensitivity (Scheiner 1993). The fact that these plasticity QTL were colocalized with QTL of traits at a certain environment may suggest that the QTL contains loci/alleles that are activated when the population is in a different environment or in an unusual condition (Paaby and Rockman 2014) and can point to the co-evolution of traits plasticity and traits at the given environment.
Supplementary information
Acknowledgements
The authors thank Simone Ariens for measurements on the microscopy images and Miriam Rodriguez for technical assistance. We thank Anne Morbach (Schlaugemacht.net) for making Fig. 1A. We thank Harm Nijveen for assistance with WormQTL2. We thank Lisa van Sluijs for the critical review of the draft manuscript. We thank Fred van Eeuwijk for helpful discussions.
Author contributions
MGS, BLS, and JEK conceived and designed the experiments. MGS and JAGR conducted the experiments. MIM analyzed the data with input from MGS. MIM and MGS wrote the manuscript, with input from JEK and BLS. All authors commented on the manuscript.
Funding
MGS was supported by NWO domain Applied and Engineering Sciences VENI grant (17282).
Data availability
The underlying data are included in the paper and interactively accessible via WormQTL2.
Materials availability
The strains used in this study can be requested from the authors.
Competing interests
The authors declare no competing interests.
Footnotes
Associate editor: Rowan Barrett.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jan E. Kammenga, Email: Jan.kammenga@wur.nl
Mark G. Sterken, Email: Mark.sterken@wur.nl
Supplementary information
The online version contains supplementary material available at 10.1038/s41437-022-00528-y.
References
- Andersen EC, Bloom JS, Gerke JP, Kruglyak L (2014) A variant in the neuropeptide receptor npr-1 is a major determinant of Caenorhabditis elegans growth and physiology. PLoS Genet 10(2):e1004156. 10.1371/journal.pgen.1004156 [DOI] [PMC free article] [PubMed]
- Andersen EC, Shimko TC, Crissman JR, Ghosh R, Bloom JS, Seidel HS, et al. A powerful new quantitative genetics platform, combining caenorhabditis elegans high-throughput fitness assays with a large collection of recombinant strains. G3. 2015;5(5):911–920. doi: 10.1534/g3.115.017178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angilletta MJ, Dunham AE. The temperature-size rule in ectotherms: simple evolutionary explanations may not be general. Am Naturalist. 2003;162:3. doi: 10.1086/377187. [DOI] [PubMed] [Google Scholar]
- Atkinson D (1994) Temperature and organism size–a biological law for ectotherms? Adv Ecol Res 25:1–58
- Azevedo RBR, French V, Partridge L. Temperature modulates epidermal cell size in Drosophila melanogaster. J Insect Physiol. 2002;48:231–237. doi: 10.1016/S0022-1910(01)00168-8. [DOI] [PubMed] [Google Scholar]
- Azevedo RBR, James AC, McCabe J, Partridge L. Latitudinal variation of wing: thorax size and wing-aspect ration in Drosophila melanogaster. Evolution. 1998;52(5):1353–1362. doi: 10.1111/j.1558-5646.1998.tb02017.x. [DOI] [PubMed] [Google Scholar]
- Bates D, Mächler M, Bolker BM, Walker SC (2015) Fitting linear mixed-effects models using lme4. J Stat Softw 67(1):1–48. 10.18637/jss.v067.i01
- Beldade P, Mateus ARA, Keller RA. Evolution and molecular mechanisms of adaptive developmental plasticity. Mol Ecol. 2011;20:1347–1363. doi: 10.1111/j.1365-294X.2011.05016.x. [DOI] [PubMed] [Google Scholar]
- Bochdanovits Z, Van Der Klis H, De Jong G. Covariation of larval gene expression and adult body size in natural populations of Drosophila melanogaster. Mol Biol Evolution. 2003;20(11):1760–1766. doi: 10.1093/molbev/msg179. [DOI] [PubMed] [Google Scholar]
- Brenner S. Genetics of the Caenorhabditis elegans. ChemBioChem. 1974;4(8):683–687. doi: 10.1002/cbic.200300625. [DOI] [Google Scholar]
- Callahan HS, Dhanoolal N, Ungerer MC. Plasticity genes and plasticity costs: a new approach using an Arabidopsis recombinant inbred population. N Phytologist. 2005;166(1):129–140. doi: 10.1111/j.1469-8137.2005.01368.x. [DOI] [PubMed] [Google Scholar]
- Carta D, Villanova L, Costacurta S, Patelli A, Poli I, Vezzù S, et al. Method for optimizing coating properties based on an evolutionary algorithm approach. Anal Chem. 2011;83(16):6373–6380. doi: 10.1021/ac201337e. [DOI] [PubMed] [Google Scholar]
- Czarnoleski M, Kramarz P, Malek D, Drobniak SM. Genetic components in a thermal developmental plasticity of the beetle Tribolium castaneum. J Therm Biol. 2017;68:55–62. doi: 10.1016/j.jtherbio.2017.01.015. [DOI] [PubMed] [Google Scholar]
- Dupuis J, Siegmund D. Statistical methods for mapping quantitative trait loci from a dense set of markers. Genetics. 1999;151(1):373–386. doi: 10.1093/genetics/151.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellers J, Driessen G. Genetic correlation between temperature-induced plasticity of life-history traits in a soil arthropod. Evolut Ecol. 2011;25:473–484. doi: 10.1007/s10682-010-9414-1. [DOI] [Google Scholar]
- Fischer K, Bauerfeind SS, Fiedler K. Temperature-mediated plasticity in egg and body size in egg size-selected lines of a butterfly. J Therm Biol. 2006;31:347–354. doi: 10.1016/j.jtherbio.2006.01.006. [DOI] [Google Scholar]
- Gaertner BE, Phillips PC. Caenorhabditis elegans as a platform for molecular quantitative genetics and the systems biology of natural variation. Genet Res. 2010;92(5–6):331–348. doi: 10.1017/S0016672310000601. [DOI] [PubMed] [Google Scholar]
- Gao AW, Sterken MG, uit de Bos J, van Creij J, Kamble R, Snoek BL, et al. Natural genetic variation in C. elegans identified genomic loci controlling metabolite levels. Genome Res. 2018;28(9):1296–1308. doi: 10.1101/gr.232322.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh SM, Testa ND, Shingleton AW (2013) Temperature-size rule is mediated by thermal plasticity of critical size in Drosophila melanogaster. Proc Biol Sci 280(1760):20130174. 10.1098/rspb.2013.0174 [DOI] [PMC free article] [PubMed]
- Gutteling EW, Doroszuk A, Riksen JAG, Prokop Z, Reszka J, Kammenga JE. Environmental influence on the genetic correlations between life-history traits in Caenorhabditis elegans. Heredity. 2007;98:206–213. doi: 10.1038/sj.hdy.6800929. [DOI] [PubMed] [Google Scholar]
- Gutteling EW, Riksen JAG, Bakker J, Kammenga JE. Mapping phenotypic plasticity and genotype – environment interactions affecting life-history traits in Caenorhabditis elegans. Heredity. 2007;98:28–37. doi: 10.1038/sj.hdy.6800894. [DOI] [PubMed] [Google Scholar]
- Jovic K, Sterken MG, Grilli J, Bevers RPJ, Rodriguez M, Riksen JAG, et al. (2017) Temporal dynamics of gene expression in heat-stressed Caenorhabditis elegans. PLoS One 12:e0189445 [DOI] [PMC free article] [PubMed]
- Kammenga JE, Doroszuk A, Riksen JAG, Hazendonk E, Spiridon L, Petrescu AJ, et al. A Caenorhabditis elegans wild type defies the temperature-size rule owing to a single nucleotide polymorphism in tra-3. PLoS Genet. 2007;3(3):0358–0366. doi: 10.1371/journal.pgen.0030034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang MH, Zaitlen NA, Wade CM, Kirby A, Heckerman D, Daly MJ, et al. Efficient control of population structure in model organism association mapping. Genetics. 2008;178(3):1709–1723. doi: 10.1534/genetics.107.080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klok CJ, Harrison JF. The temperature size rule in Arthropods: independent of macro-environmental variables but size dependent. Integr Comp Biol. 2013;53(4):557–570. doi: 10.1093/icb/ict075. [DOI] [PubMed] [Google Scholar]
- Kruijer W, Boer MP, Malosetti M, Flood PJ, Engel B, Kooke R, et al. Marker-based estimation of heritability in immortal populations. Genetics. 2014;199(2):379–398. doi: 10.1534/genetics.114.167916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafuente E, Beldade P. Genomics of developmental plasticity in animals. Front Genet. 2019;10:1–18. doi: 10.3389/fgene.2019.00720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafuente E, Duneau D, Beldade P. Genetic basis of thermal plasticity variation in Drosophila melanogaster body size. PLoS Genet. 2018;14(9):1–24. doi: 10.1371/journal.pgen.1007686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Álvarez OA, Gutteling EW, Tijsterman M, Fu J, Riksen JAG, et al. Mapping determinants of gene expression plasticity by genetical genomics in C. elegans. PLoS Genet. 2006;2(12):2155–2161. doi: 10.1371/journal.pgen.0020222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima T, Ishiura S, Suo S. Regulation of body size in Caenorhabditis elegans: effects of environmental factors and the nervous system. Int J Developmental Biol. 2017;61(6–7):367–374. doi: 10.1387/ijdb.160352ss. [DOI] [PubMed] [Google Scholar]
- Nakad R, Snoek LB, Yang W, Ellendt S, Schneider F, Mohr TG, et al. Contrasting invertebrate immune defense behaviors caused by a single gene, the Caenorhabditis elegans neuropeptide receptor gene npr-1. BMC Genomics. 2016;17(1):1–20. doi: 10.1186/s12864-016-2603-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norry FM, Loeschcke VR. Longevity and resistance to cold stress in cold-stress selected lines and their controls in Drosophila melanogaster. J Evolut Biol. 2002;15:775–783. doi: 10.1046/j.1420-9101.2002.00438.x. [DOI] [Google Scholar]
- Paaby AB, Rockman MV. Cryptic genetic variation: evolution’ s hidden substrate. Nat Rev Genet. 2014;15(4):247–258. doi: 10.1038/nrg3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng IF, Berke BA, Zhu Y, Lee WH, Chen W, Wu CF. Temperature-dependent developmental plasticity of drosophila neurons: cell-autonomous roles of membrane excitability, Ca2+ influx, and cAMP signaling. J Neurosci. 2007;27(46):12611–12622. doi: 10.1523/JNEUROSCI.2179-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen C, Dirksen P, Schulenburg H. Why we need more ecology for genetic models such as C. elegans. Trends Genet. 2015;31(3):120–127. doi: 10.1016/j.tig.2014.12.001. [DOI] [PubMed] [Google Scholar]
- R Core Team (2021) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
- Reddy KC, Andersen EC, Kruglyak L, Kim DH. A polymorphism in npr-1 is a behavioral determinant of pathogen susceptibility in C. elegans. Science. 2009;323(5912):382–384. doi: 10.1126/science.1166527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg LH, Archer MA, Wayne RK (1999) Transgressive segregation, adaptation and speciation. Heredity 83:363–372 [DOI] [PubMed]
- Rockman MV, Skrovanek SM, Kruglyak L (2010) Selection at linked sites shapes. Science 330:372–376. 10.1126/science.1194208 [DOI] [PMC free article] [PubMed]
- Rodriguez M, Snoek LB, Riksen JAG, Bevers RP, Kammenga JE. Genetic variation for stress-response hormesis in C. elegans lifespan. Exp Gerontol. 2012;47(8):581–587. doi: 10.1016/j.exger.2012.05.005. [DOI] [PubMed] [Google Scholar]
- Saltz JB, Bell AM, Flint J, Gomulkiewicz R, Hughes KA, Keagy J. Why does the magnitude of genotype-by-environment interaction vary? Ecol Evolution. 2018;8(12):6342–6353. doi: 10.1002/ece3.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiner S. Plasticity as a selectable trait: reply to via. Am Soc Naturalist. 1993;142(2):371–373. doi: 10.1086/285544. [DOI] [Google Scholar]
- Sgro C, Hoffmann A (2004) Genetic correlations, tradeoffs and environmental variation. Heredity 93:241–248. 10.1038/sj.hdy.6800532 [DOI] [PubMed]
- Snoek BL, Sterken MG, Bevers RPJ, Volkers RJM, van Hof A, Brenchley R, et al. (2017) Contribution of trans regulatory eQTL to cryptic genetic variation in C. elegans. BMC Genomics 18:500. 10.1186/s12864-017-3899-8 [DOI] [PMC free article] [PubMed]
- Snoek BL, Volkers RJM, Nijveen H, Petersen C, Dirksen P, Sterken MG, et al. (2019) A multi-parent recombinant inbred line population of C. elegans allows identification of novel QTLs for complex life history traits. BMC Biol 17:24 [DOI] [PMC free article] [PubMed]
- Snoek LB, Orbidans HE, Stastna JJ, Aartse A, Rodriguez M, Riksen JAG, et al. Widespread genomic incompatibilities in Caenorhabditis elegans. G3: Genes, Genomes, Genet. 2014;4(10):1813–1823. doi: 10.1534/g3.114.013151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoek LB, Sterken MG, Hartanto M, van Zuilichem AJ, Kammenga JE, de Ridder D et al. (2020) WormQTL2: an interactive platform for systems genetics in Caenorhabditis elegans. Database 2020:baz149. 10.1093/database/baz149 [DOI] [PMC free article] [PubMed]
- Steigenga MJ, Zwaan BJ, Brakefield PM, Fischer K. The evolutionary genetics of egg size plasticity in a butterfly. J Evolut Biol. 2005;18:281–289. doi: 10.1111/j.1420-9101.2004.00855.x. [DOI] [PubMed] [Google Scholar]
- Sterken MG, Bevers RPJ, Volkers RJM, Riksen JAG, Kammenga JE, Snoek BL. Dissecting the eQTL micro-architecture in Caenorhabditis elegans. Front Genet. 2020;11(Nov):1–15. doi: 10.3389/fgene.2020.501376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterken MG, Plaat LVB, Van Der Riksen JAG, Rodriguez M, Schmid T, Hajnal A, et al. (2017) Ras/MAPK modifier loci revealed by eQTL in Caenorhabditis elegans. G3 (Bethesda) 7:3185–3193. 10.1534/g3.117.1120 [DOI] [PMC free article] [PubMed]
- Sterken MG, Snoek LB, Kammenga JE, Andersen EC. The laboratory domestication of Caenorhabditis elegans. Trends Genet. 2015;31(5):224–231. doi: 10.1016/j.tig.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Têtard-jones C, Kertesz M, Preziosi R. Quantitative trait loci mapping of phenotypic plasticity and genotype-environment interactions in plant and insect performance. Philos Trans R Soc B: Biol Sci. 2011;366:1569. doi: 10.1098/rstb.2010.0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson OA, Snoek LB, Nijveen H, Sterken MG, Volkers RJM, Brenchley R, et al. Remarkably divergent regions punctuate the genome assembly of the Caenorhabditis elegans Hawaiian strain CB4856. Genetics. 2015;200(3):975–989. doi: 10.1534/genetics.115.175950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Voorhies WA. Bergmann size clines: a simple explanation for their occurrence in ectotherms. Evolution. 1996;50(3):1259–1264. doi: 10.1111/j.1558-5646.1996.tb02366.x. [DOI] [PubMed] [Google Scholar]
- Via S, Gomulkiewicz R, de Jong G, Scheiner SM, Schlichting CD, van Tienderen PH. Adaptive phenotypic plasticity: consensus and controversy. Trends Ecol Evolution. 1995;10:5. doi: 10.1016/S0169-5347(00)89061-8. [DOI] [PubMed] [Google Scholar]
- Viñuela A, Snoek LB, Riksen JAG, Kammenga JE. Genome-wide gene expression regulation as a function of genotype and age in C. elegans. Genome Res. 2010;20(7):929–937. doi: 10.1101/gr.102160.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viñuela A, Snoek LB, Riksen JAG, Kammenga JE (2011) Gene expression modifications by temperature-toxicants interactions in Caenorhabditis elegans. PLoS One 6(9):e24676. 10.1371/journal.pone.0024676 [DOI] [PMC free article] [PubMed]
- Wickham H. Ggplot2. Wiley Interdiscip Rev: Computational Stat. 2011;3(2):180–185. doi: 10.1002/wics.147. [DOI] [Google Scholar]
- Wickham H, Averick M, Bryan J, Chang W, McGowan L, François R, et al. Welcome to the Tidyverse. J Open Source Softw. 2019;4(43):1686. doi: 10.21105/joss.01686. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The underlying data are included in the paper and interactively accessible via WormQTL2.
The strains used in this study can be requested from the authors.