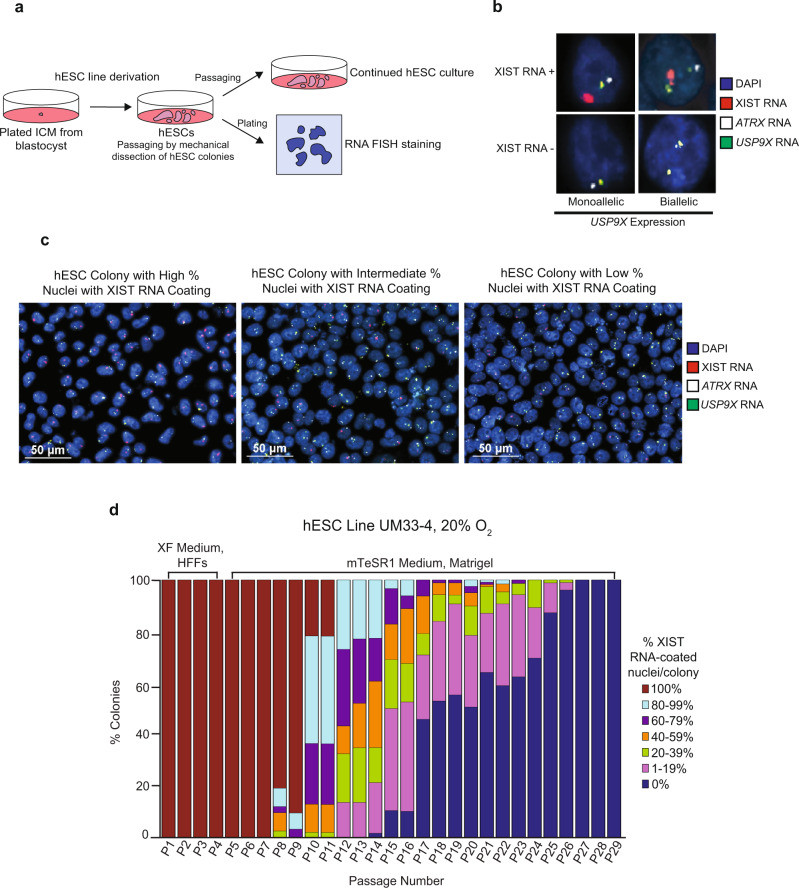
Fig. 1. Loss of XIST RNA coating upon prolonged passaging of female hESCs.
a Schematic depicting the derivation, culture, passaging, and RNA FISH staining of hESCs in this study. b Representative nuclei stained to detect XIST RNA (red), RNAs from X-linked genes ATRX (white) and USP9X (green), and the nucleus with DAPI (blue). Top, representative nuclei with XIST RNA coating. Bottom, representative nuclei without XIST RNA coating. At least 100 nuclei were counted per colony for hESC RNA FISH quantification. The total number of colonies quantified at each passage range from 2–138 and are cataloged in source data. c Representative hESC colonies stained to detect XIST RNA (red), ATRX RNA (white), and USP9X RNA (green). Nuclei are stained blue with DAPI. At least 100 nuclei were counted per colony for hESC RNA FISH quantification. d Percentage of nuclei with XIST RNA coats per colony in hESC line UM33-4 derived in XenoFree (XF) medium on human fibroblast feeders (HFFs) and cultured subsequently on Matrigel under atmospheric oxygen levels (20%). The percentage of nuclei with XIST RNA coats in individual hESC colonies were stratified into 20% increments. 100% value indicates that all nuclei in a colony harbored XIST RNA coats, whereas 0% indicates that all nuclei lacked XIST RNA coats in a colony. The percentage of colonies harboring nuclei with XIST RNA coats decreased significantly with passage number (general linear model, p = 0.002). See also Supplementary Fig. 2. Source data are provided as a Source Data file.