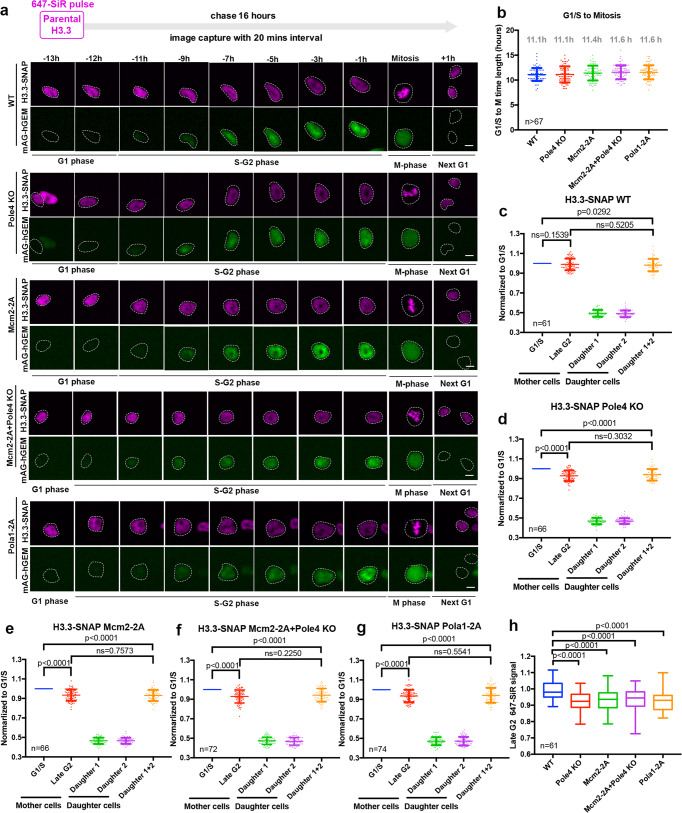
Fig. 4. Pola1 facilitates parental histone H3.3 recycling.
a Representative live-cell images of parental histones H3.3-SNAP signals at the indicated time points in WT (n = 61), Pole4 KO (n = 66), Mcm2-2A (n = 66), Pola1-2A (n = 74) mES single and Mcm2-2A Pole4 KO double mutant (n = 72) cell lines. Purple channels are 647-SiR labeled-parental H3.3-SNAP, green channels are for mAG tagged-geminin expression. Scale bar, 10 µm. Tracked cells were circled. b The average time from G1/S to mitosis for WT, Pole4 KO, Mcm2-2A, Pola1-2A, and Mcm2-2A Pole4 KO double mutant mES cell lines based on the appearance of mAG tagged-geminin (G1/S) and the reduction of mAG tagged-geminin signals (mitosis). c–g Quantification of H3.3-SNAP signals at G1/S and G2 of each mother cell and G1 of its two daughter cells in WT, Pole4 KO, Mcm2-2A, Pola1-2A, and Mcm2-2A + Pole4 KO mutant mES cell lines. The cell cycle stage of each cell was based on mAG tagged-geminin signals. h Comparison of the relative amount of parental H3.3-SNAP at individual cells at late G2 in WT, Pole4 KO, Mcm2-2A, Pola1-2A and Mcm2-2A + Pole4 KO mES cell lines. The center line, the box limits and the whiskers were defined as described in Fig. 2c. b–g Data are presented as means ± SD. a–h n number of cells from two independent experiments. c–h Statistical analysis was performed by two-tailed unpaired Student t test with the P values marked on the graphs, ns no significant difference.