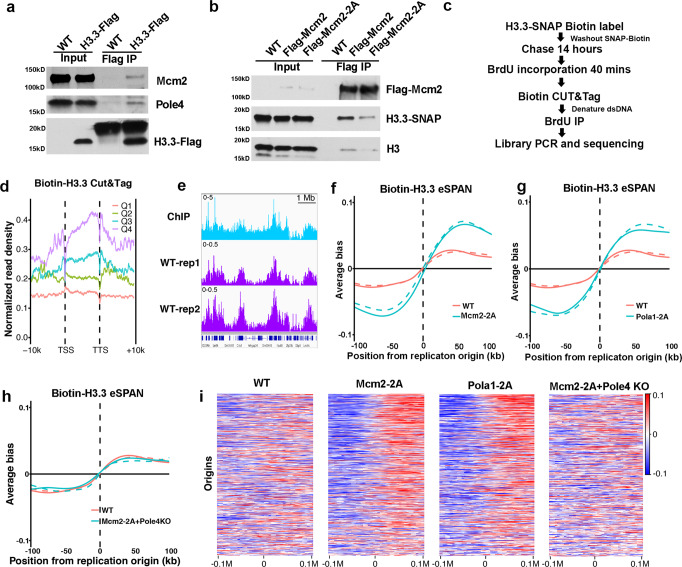
Fig. 5. Mcm2 and Pole4 interact with H3.3 and facilitate parental H3.3 transfer to different replicating DNA strands.
a Both Mcm2 and Pole4 interact with H3.3 in vivo. WT or H3.3-Flag-tagged mES cells were collected for immunoprecipitation using anti-Flag antibodies. Proteins in the input extracts and IP samples were analyzed by Western blotting using Flag, Mcm2 and Pole4 antibodies. One representative result from three independent replicates was shown. b Mcm2-2A mutation reduces the Mcm2-H3.3 interaction. Cell extracts from WT, Mcm2-Flag, Mcm2-2A-Flag mouse cells containing H3.3-SNAP were used for immunoprecipitation using anti-Flag antibodies. Proteins in the input and IP samples were analyzed by Western blot using Flag, SNAP and H3 antibodies. One representative results from three independent replicates was shown. c An outline of the experimental procedures to analyze the distribution of parental H3.3-SNAP at replicating DNA using eSPAN. d Parental H3.3-SNAP CUT&Tag density at TSS and TTS at genes with different expression. Genes were separated into 4 groups based on their expression in mouse ES cells (Q1 = lowest, Q4 = highest). H3.3-SNAP CUT&Tag density was calculated for each group. e Snapshots of H3.3 ChIP-Seq (GSM2080326) and two repeats of H3.3-SNAP CUT&Tag density at selected region (chr5:99,655,789–106,254,109) in wild-type cells. The signals represent the normalized read count per million reads for each of the three indicated setting. f The average bias of parental H3.3-eSPAN peaks at 1548 replication origins in WT and Mcm2-2A mES cells. The eSPAN bias at each origin was calculated using the formula (W − C)/(W + C); W and C: sequence reads of the Watson strand and Crick strand, respectively. Two repeats are shown. g, h The average bias of parental H3.3-eSPAN peaks at 1548 replication origins in WT and Pola1-2A mES cells (g) as well as WT and Mcm2-2A Pole4 KO double mutant mES cells (h). Two repeats are shown. i Heatmaps of parental H3.3-SNAP eSPAN bias in WT, Mcm2-2A, Pola1-2A and Mcm2-2A + Pole4 KO mouse ES cells at each of the 1548 initiation zones, ranked from the most efficient (top) to the least efficient (bottom) ones based on OK-seq bias.