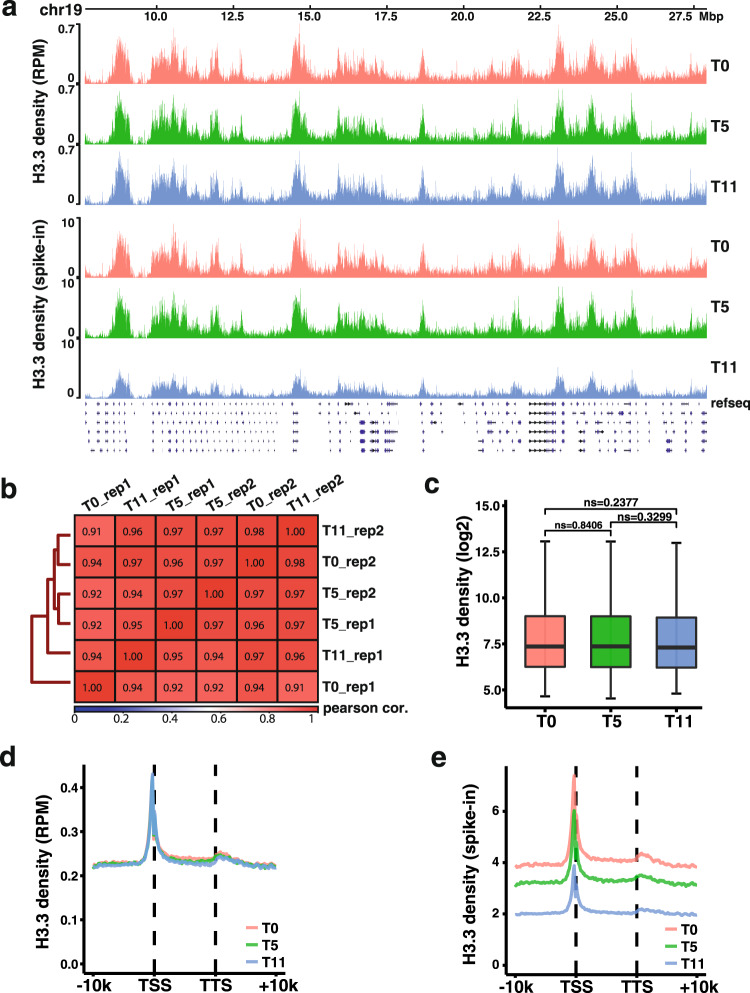
Fig. 6. Parental H3.3 proteins are likely recycled at chromatin regions locally.
a Snapshot of Parental H3.3-SNAP CUT&RUN density at chromosome 19 (Chr19) at different three time points, 0 h (T0), 5 h (T5), and 11 h (T11) after release into fresh media without SNAP-biotin substrate in WT mouse ES cells. Reads were normalized either against total mapped reads at each time (top panels) or DNA from spike-in human HeLa cells at each time point (lower panels). b Pairwise Pearson correlation matrix of two biological replicates of parental H3.3-SNAP CUT&RUN signals at each time point across the whole genome using a 50 kb window. c The average of parental H3.3-SNAP CUT&RUN density at different time points (T0, T5, and T11) on chromatin regions enriched with H3.3 after normalizing to total mapped reads at each time point. Average of two biological replicates is shown. n = 2461 chromatin regions were analyzed for each time points. The center line, the box limits and the whiskers are defined as in Fig. 2c. Statistical analysis was performed by two-tailed unpaired Student t test. The P values are marked on the graphs (ns, no significant difference). d The average of H3.3-SNAP CUT&RUN density of each time point (T0, T5, and T11) at the TSS and TTS of 5271 genes localized at replicated chromatin regions after normalized with total mapped reads. Average of two biological replicates is shown. e H3.3-SNAP CUT&RUN density of each time point (T0, T5, and T11) at the TSS and TTS of 5271 genes localized at replicating chromatin when normalized against DNA from spike-in HeLa cells. The average of two biological replicates is shown.