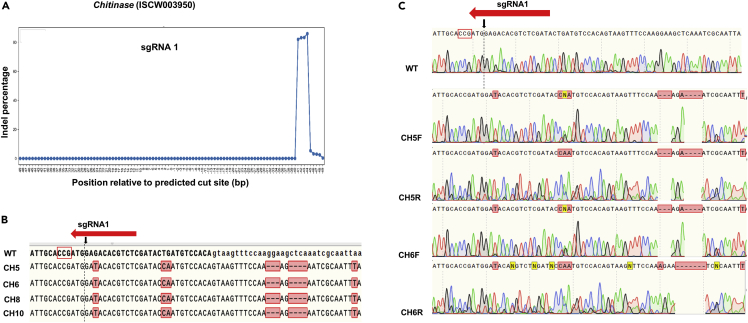
Figure 3.
Sequences of Chitinase G0 larvae edited by embryo injection aligned to wildtype (WT)
(A) Summary of deep sequencing data of animals injected with sgRNA 1. Indel percentage is on the Y axis. The X axis depicts predicted Cas9 cut site (position 0) and sequences up- and downstream of the cut site.
(B) Representative sequences containing deletions corresponding to sgRNA 1. The predicted cut site is shown by a black arrow and dotted line. PAM site is indicated by a red rectangle outline. Alignments were generated using Snapgene.
(C) Forward (F) and Reverse (R) chromatograms of WT, CH5, and CH6 larvae. Additional sequences, chromatograms, and indel/substitution percentages calculated by deep sequencing are shown in Figure S5 and Table S4.