FIGURE 1.
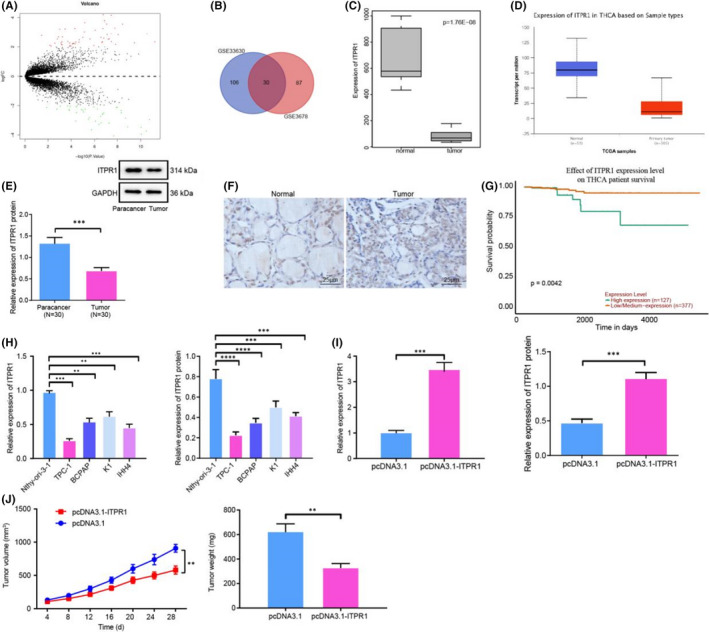
ITPR1 is poorly expressed in PTC tissues and cells and alleviates tumour growth. (A) Volcano plot of differentially expressed genes in PTC. Abscissa: logFC; Ordinate: log10 p value. A red dot refers to up‐regulated gene; a green dot refers to down‐regulated genes. (B) Venn diagram of PTC differentially expressed genes from profiling microarrays GSE33630 and GSE3678. Number refers to amount of genes. (C) Box plot of ITPR1 expression in PTC samples and normal samples from GSE3678. (D) Expression level of ITPR1 in PTC predicted by Ualcan database. (E) RT‐qPCR and Western blot analyses of ITPR1 expression in 30 paired PTC tissues and adjacent normal tissues. ***p < 0.001 versus adjacent normal tissues (F) IHC of ITPR1 expression in PTC tissues and adjacent normal tissues (400×). (G) RFS survival analysis of ITPR1 and prognosis of PTC patients. (H) RT‐qPCR and Western blot analyses of ITPR1 expression in TPC‐1, BCPAP, K1, IHH4 and Nthy‐ori‐3‐1 cells. **p < 0.01, and ***p < 0.001 versus Nthy‐ori‐3‐1 cells. (I) RT‐qPCR and Western blot analyses of ITPR1 expression level in TPC‐1 cells upon transfection of oe‐ITPR1. **p < 0.01 versus pcDNA3.1. (J) Quantification of mouse tumour after injection of oe‐ITPR1 cells and quantification of tumour volume within days after treatment. (K) Quantification of tumour weight after injection of oe‐ITPR1 cells. *p < 0.05, **p < 0.01, and ***p < 0.001 versus pcDNA3.1. Measurement data were expressed as mean ± standard deviation. The cancer tissues and adjacent normal tissues were compared by paired t‐test while analysis of the other two group was performed through unpaired t‐test. Analysis among multiple groups was conducted by ANOVA followed by Tukey's post hoc test. Data at different time points among groups were compared by repeated measures ANOVA, followed by Bonferroni
