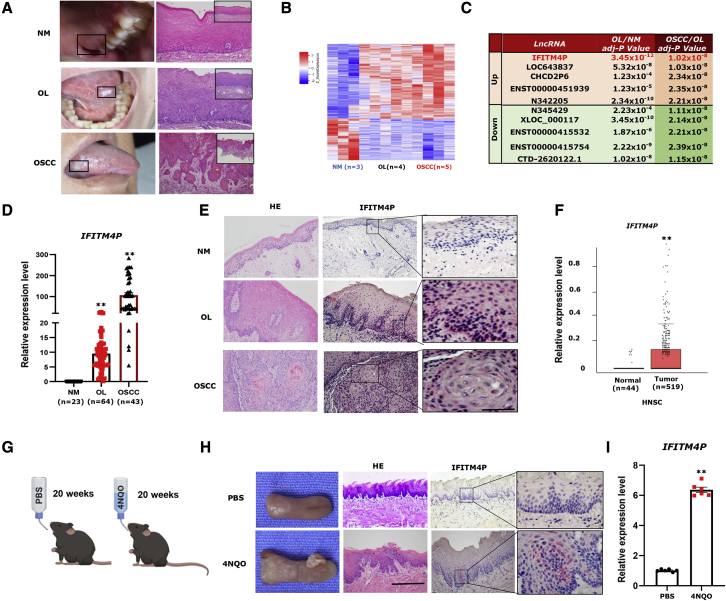
Figure 1.
IFITM4P was upregulated in OL and OSCC compared with NM
(A) Typical macroscopic and microscopic findings (H&E staining) in NM, OL, and OSCC (100×). (B) Hierarchical clustering microarray analysis was performed on NM (n = 3), OL (n = 4), and OSCC (n = 5) samples from Chinese patients based on differentially expressed RNA transcripts (p < 0.05, fold change >2) from the microarray data. Each column represents a sample and each row represents a transcript. The expression level of each gene in a single sample is depicted according to the color scale. (C) Identification of regulated lncRNAs in OL/NM and OSCC/OL and list of ten differentially expressed lncRNAs with fold change >2 and p < 0.001. (D) qRT-PCR analysis showed IFITM4P expression was highest in OSCC samples compared with OL and NM samples, with the lowest in NM samples, ∗∗p < 0.001. (E) In the stepwise samples from the same patient, IFITM4P staining became stronger during development of OSCC from OL (200×). IFITM4P staining appears negative in adjacent NM. (F) TCGA data show that higher expression of IFITM4P was seen in HNSC tissues (n = 519) than in normal tissues (n = 44), ∗∗p < 0.001. (G) Schematic timeline of the tongue leukoplakia/SCC model. (H) Typical tongue leukoplakia and SCC were found in the 4NQO group. The histopathological diagnosis also confirmed leukoplakia on the dorsal tongue and local early invasive tongue SCC in the 4NQO group. FISH showed strong IFITM4P staining in the 4NQO group, while no staining was found in the PBS group (n = 6). (I) qRT-PCR results showed that IFITM4P was increased in the 4NQO group compared with the PBS group. (n = 6), ∗∗p < 0.001.