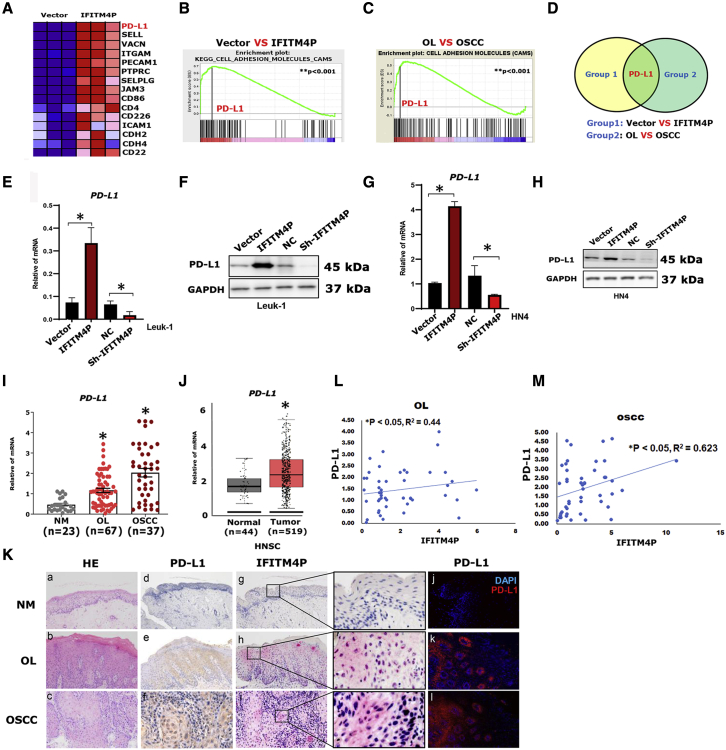
Figure 3.
PD-L1 is a novel downstream target of IFITM4P in OL and OSCC
(A) Heatmap of cell adhesion-associated genes (by GSEA analysis) in the leading edge showing the strongest upregulation in the IFITM4P group in Leuk-1 cells. (B) Comparison of the enrichment plots for vector versus IFITM4P-expressing Leuk-1 cells generated by GSEA analysis of ranked gene expression data (left, upregulated [red]; right, downregulated [blue]). The enrichment score is shown as a green line (enrichment score = 0.52; ∗∗p < 0.001). (C) Comparison of enrichment plots for OL versus OSCC generated by GSEA analysis of ranked gene expression data (left, upregulated [red]; right, downregulated [blue]). The enrichment score is shown as a green line (enrichment score = 0.49; ∗∗p < 0.001). (D) Venn diagram showing the overlap between vector versus IFITM4P-expressing Leuk-1 cells and OL versus OSCC. (E and F) Induction of PD-L1 by IFITM4P was confirmed by qRT-PCR (E) and WB (F) in Leuk-1 cells. (G and H) Induction of PD-L1 by IFITM4P was confirmed by qRT-PCR (G) and WB (H) in HN4 cells. (I) qRT-PCR analysis showed that PD-L1 expression was highest in OSCC samples compared with OL and NM samples, with the lowest in NM samples. (J) TCGA data analysis showed higher PD-L1 expression in HNSC tissues (n = 519) than in normal tissues (n = 44), ∗p < 0.05. (K) In the samples from patients, no PD-L1 and IFITM4P staining was seen in NM. However, PD-L1 and IFITM4P staining became stronger as OL progressed to early invasive OSCC (a–l, 200×). (L and M) Positive correlation between the levels of IFITM4P and PL-D1 in OL (∗p < 0.05) (L) and OSCC (∗p < 0.05) (M) samples. Data from (E), (G), and (I) are shown as mean ± SD from three independent experiments. ∗p < 0.05.