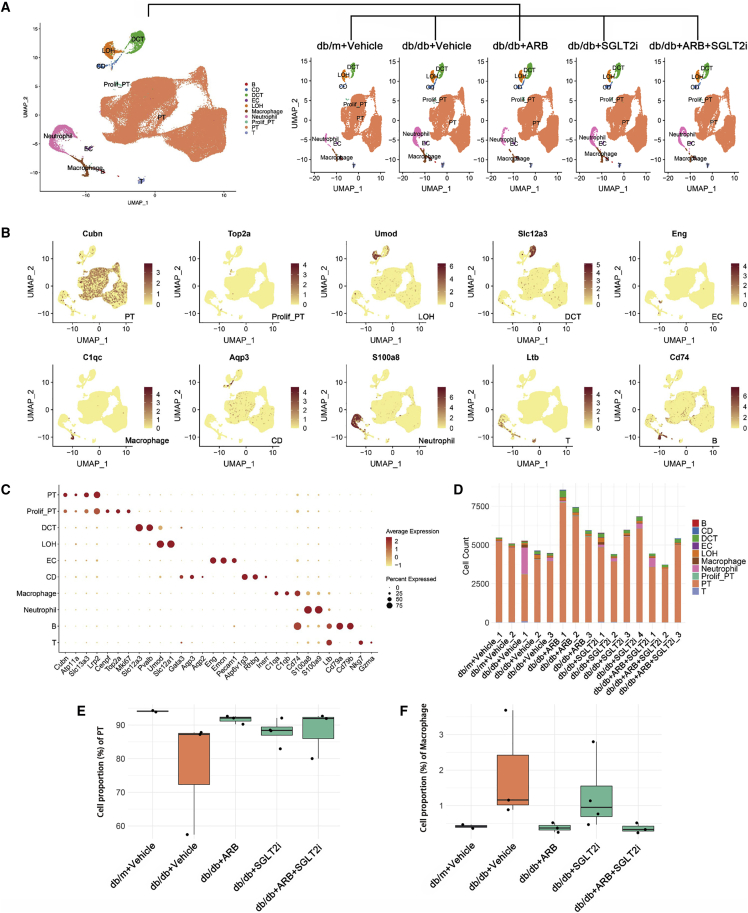
Figure 1.
Cell diversity in mouse kidney cells delineated by single-cell transcriptomic analysis
(A) All samples with different treatments were integrated into a single dataset and clustered using UMAP. Colors and labels indicate different cell types based on marker gene expression. B, B cells; CD, collecting duct; DCT, distal convoluted tubule; EC, endothelial cell; LOH, loop of henle; PT, proximal tubule; prolif-PT, proliferative PT; T, T cells. (B) Expression of selected marker genes for each cell type projected on UMAP. (C) Dot plot shows the expression of marker genes for the identified cell types. (D) Number of identified cell types in each sample. (E and F) Boxplots of the average proportion of PTs (E) and macrophages (F) in different groups.