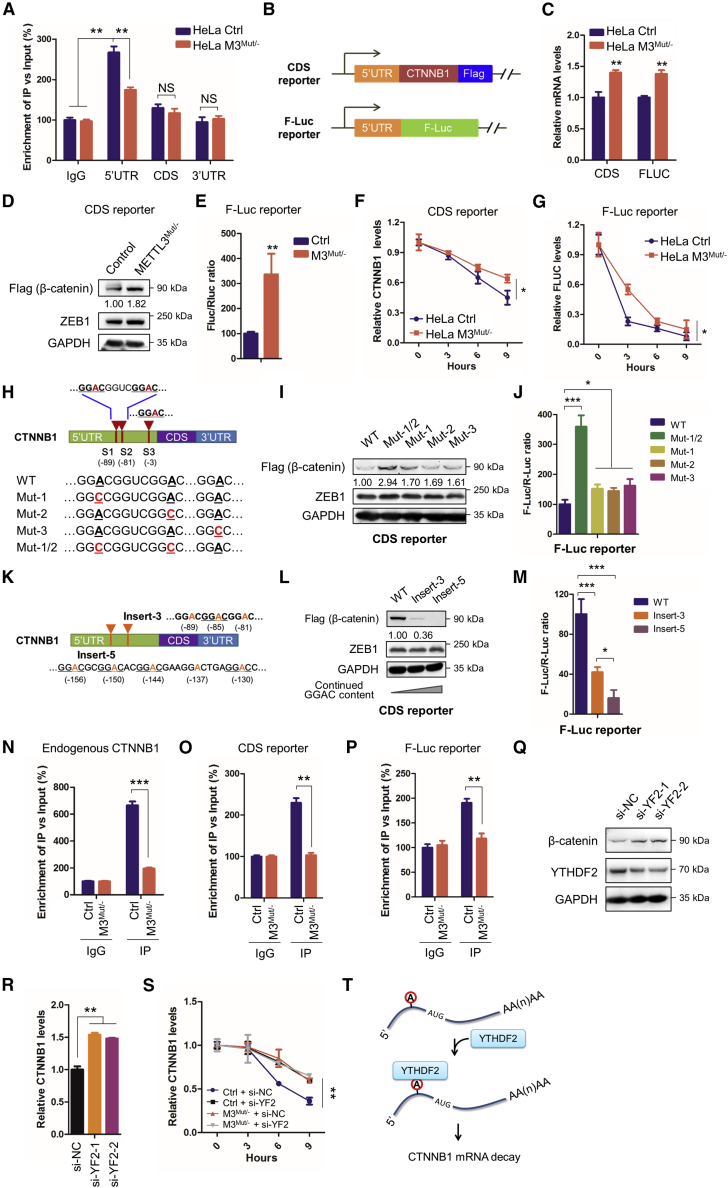
Figure 5.
5′UTR m6A methylation destabilizes CTNNB1 mRNA
(A) m6A RIP-qPCR analysis of CTNNB1 mRNA in Con or M3Mut/− HeLa cells using fragmented RNA. (B) Schematic of the CDS reporter and F-Luc reporter, which contains the CTNNB1 5′UTR before CTNNB1 CDS or reporter genes. (C) The CDS reporter and F-Luc reporter were transfected into Con and M3Mut/− HeLa cells, respectively. Expression levels of reporter mRNA were detected by quantitative real-time PCR: CTNNB1 mRNA for the CDS reporter, normalized to endogenous CTNNB1 and GAPDH mRNA levels, and F-LUC mRNA for the F-Luc reporter, normalized to R-LUC mRNA. (D) The CDS reporter was transfected into Con or M3Mut/− HeLa cells for 48 h. Expression levels of exogenous β-catenin (FLAG) were detected by western blot. Band intensities of β-catenin were analyzed using ImageJ and are listed at the bottom of the target bands. (E) The F-Luc reporter was transfected into Con or M3Mut/− HeLa cells for 48 h. A dual-luciferase assay was performed to measure F-Luc production, which was normalized to R-Luc levels. (F) The CDS reporter was transfected into Con or M3Mut/− HeLa cells for 48 h and then treated with Act-D for the indicated times. The expression levels of CTNNB1 mRNA were detected by quantitative real-time PCR. (G) The F-Luc reporter was transfected into Con or M3Mut/− HeLa cells for 48 h and then treated with Act-D for the indicated times. Expression levels of F-LUC mRNA were detected by quantitative real-time PCR. (H) Schematic of m6A sites within CTNNB1 mRNA. Mutations of m6A sites in the CTNNB1 5′UTR are marked in red. (I) WT and mutated CDS reporters were transfected into HeLa cells. Expression levels of exogenous β-catenin (FLAG) were detected by western blot. Band intensities of FLAG-β-catenin were analyzed using ImageJ and are listed at the bottom of the target bands. (J) WT and mutated F-Luc reporters were transfected into HeLa cells. A dual-luciferase assay was performed to measure F-Luc production, which was normalized to R-Luc levels. (K) Schematic of m6A insertions within CTNNB1 mRNA. Insertions of continued m6A motifs in the CTNNB1 5′UTR are marked in orange. (L) WT and CDS reporters with inserted m6A motifs were transfected into HeLa cells. Expression levels of exogenous β-catenin (FLAG) were detected by western blot. Band intensities of FLAG-β-catenin were analyzed using ImageJ and are listed at the bottom of the target bands. (M) WT and F-Luc reporters with inserted GGAC motifs were transfected into HeLa cells. A dual-luciferase assay was performed to measure F-Luc production, which was normalized to R-Luc levels. (N) YTHDF2-RIP analysis of CTNNB1 mRNA in Con and M3Mut/− HeLa cells. (O) CDS reporters were transfected into Con and M3Mut/− HeLa cells. YTHDF2 RIP analysis of reporter mRNA was performed. (P) F-Luc reporters were transfected into Con and M3Mut/− HeLa cells. YTHDF2-RIP analysis of reporter mRNA was performed. (Q) Expression levels of β-catenin in HeLa cells silencing YTHDF2 were detected by western blot. (R) Expression levels of CTNNB1 mRNA in HeLa cells silencing YTHDF2 were detected by quantitative real-time PCR, (S) Con or M3Mut/−- HeLa cells were transfected with si-NC or si-YTHDF2 and then treated with CHX for the indicated time. Expression of CTNNB1 mRNA was detected by quantitative real-time PCR. (T) Mechanism of m6A regulating the stability of CTNNB1 mRNA. Data are presented as mean ± SD from three independent experiments. Student’s t test; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.