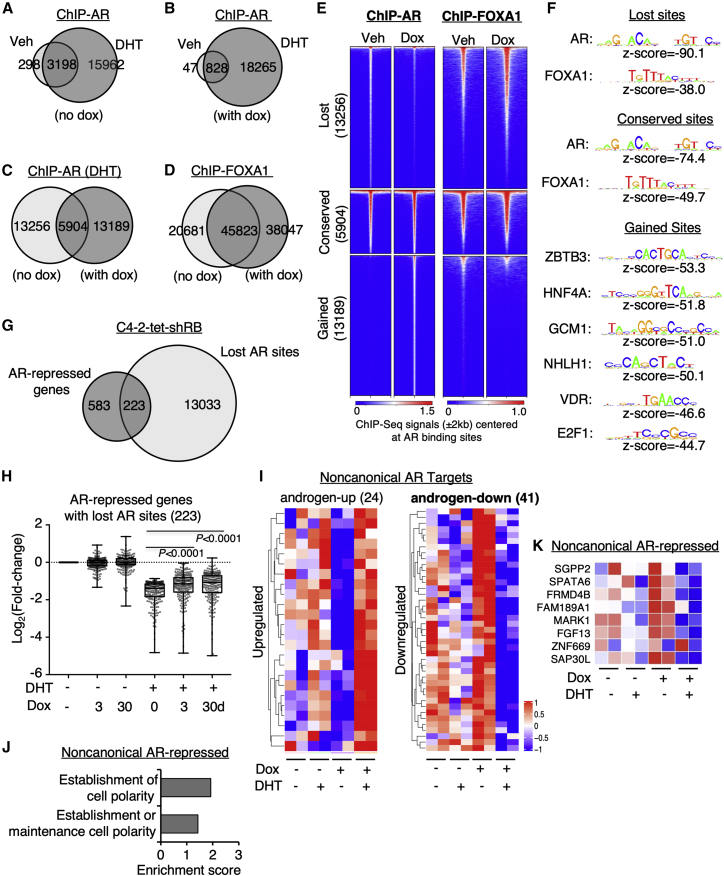
Figure 4.
Rb depletion reprograms the transcriptional repression activity of AR
(A–C) ChIP-seq analyses of AR were performed in C4-2-tet-shRB cells cultured with vehicle or doxycycline (3 days) and then stimulated with ethanol or DHT (10 nM, 4 h). Venn diagrams for AR binding peaks in cells treated with DHT versus ethanol in the absence of doxycycline (A), DHT versus ethanol in the presence of doxycycline (B), and doxycycline versus vehicle in the presence of DHT (C) were shown. (D) ChIP-seq analyses of FOXA1 were performed in C4-2-tet-shRB cells cultured with vehicle or doxycycline. The Venn diagram for FOXA1 binding peaks in both conditions was shown. (E) Heatmap view for ChIP-seq signal intensity of AR and FOXA1 at clustered AR-binding sites (lost, conserved, and gained) is shown. (F) Motif enrichment analysis for these AR binding sites is shown. (G) The Venn diagram for AR-repressed genes and the gene annotation of the “lost” AR sites is shown. (H) Box plots for the change of expression of AR-repressed genes containing at least one lost AR site in C4-2-tet-shRB cells are shown. (I) Heatmap view for RB1-silencing-induced AR-reprogramming targets determined by gained AR binding and regulation (fold-change > 1.5) is shown. (J) Gene ontology analyses for the reprogrammed AR-repressed genes are shown. (K) Heatmap view for a panel of reprogrammed AR-repressed genes is shown.