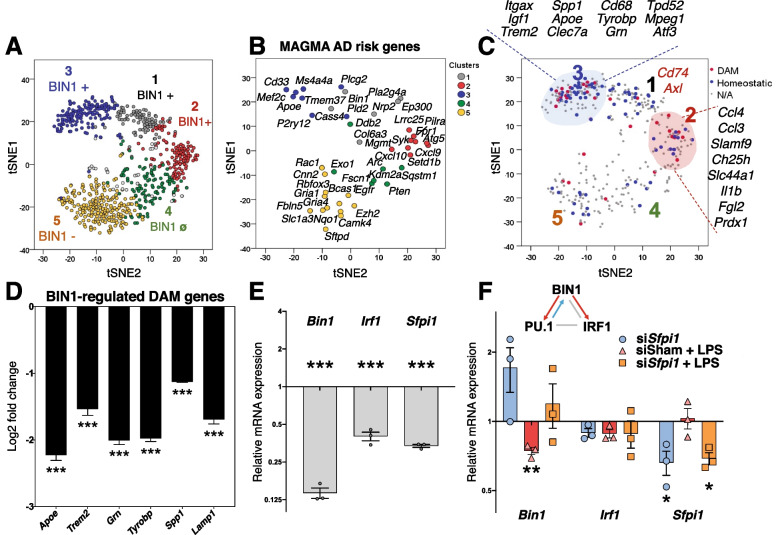
Fig. 3.
Genes affected by Bin1 KD in vitro are implicated in AD and regulation of microglial phenotypes. A Visualisation of in vitro microglial transcriptomic data using t-SNE shows gene clusters positively (clusters 1-3) or negatively regulated by BIN1 (cluster 5). See the heatmap in Fig. 2D for the cluster color reference. One cluster was unaffected by BIN1 expression (cluster 4). B MAGMA of AD-associated risk genes overlapped with our dataset, demonstrating crucial AD-related genes within each cluster that are regulated downstream of microglial BIN1. C Critical disease-associated (DAM shown in red) and homeostatic microglial genes (shown in blue) were dysregulated by Bin1 KD in primary microglia. DAM and homeostatic assignments were based on published literature [25]. D qRT-PCR validation confirmed that BIN1 positively regulates several key DAM genes, including Apoe, Trem2, and Tyrobp (i.e., down-regulated by Bin1 KD) (*p < 0.05, **p < 0.01, ***p < 0.001, two-tailed t-test comparing sham siRNA to Bin1 siRNA conditions, normalized to Gapdh, n = 3/condition). E Bin1 KD causes down-regulation of two master transcriptional regulators of microglial phenotypes – Irf1 and Sfpi1 (encoding PU.1). (F) The plot depicts the q-PCR analysis of the relative change in Bin1, Irf1, and Sfpi1 transcript abundance compared to the sham siRNA condition. siRNA KD of Sfpi1 demonstrates co-dependent regulation between BIN1 and PU.1