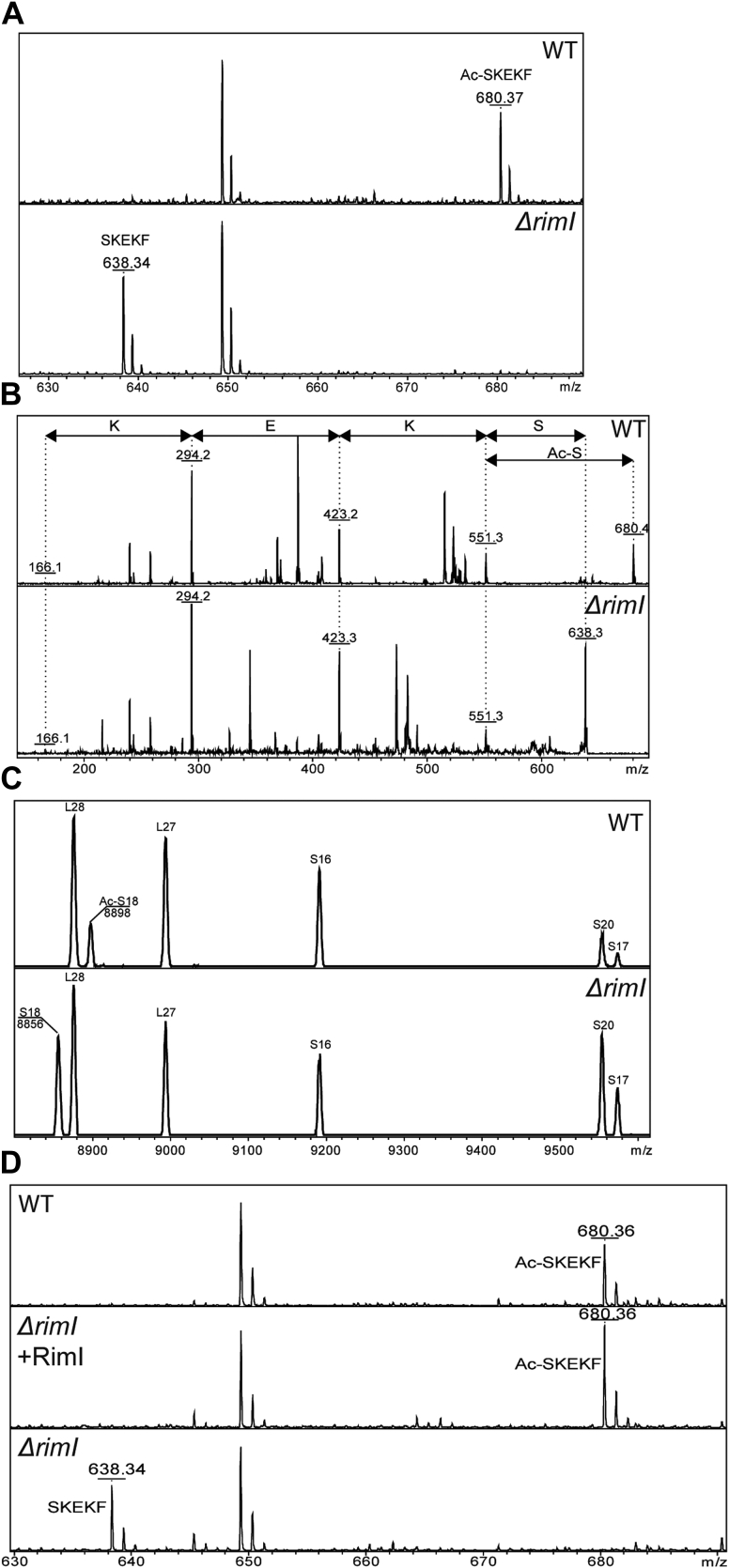
Figure 2.
Mass spectrometry analysis of protein modification in the WT and ΔrimI strain (panels are labeled accordingly).A, mass spectrometry analysis of EF-Tu modification. EF-Tu hydrolysate by chymotrypsin was analyzed. Panels corresponding to the WT and ΔrimI strain are labeled. Shown are peaks corresponding to the N-terminal SKEKF peptide. B, fragmentation analysis of the N-terminal peptide of EF-Tu. Panels corresponding to the WT and ΔrimI strain are labeled. Mass differences between the fragments are labeled by amino acids removed. C, mass spectrometry analysis of S18 protein. Total ribosomal proteins were used for the analysis. The panels correspond to the WT strain (upper panel) and the ΔrimI strain (lower panel), labeled accordingly. Peaks corresponding to ribosomal proteins are marked. D, genetic complementation of the ΔrimI strain by the plasmid coding for RimI. Shown are peaks corresponding to the N-terminal SKEKF peptide. EF-Tu, elongation factor Tu.