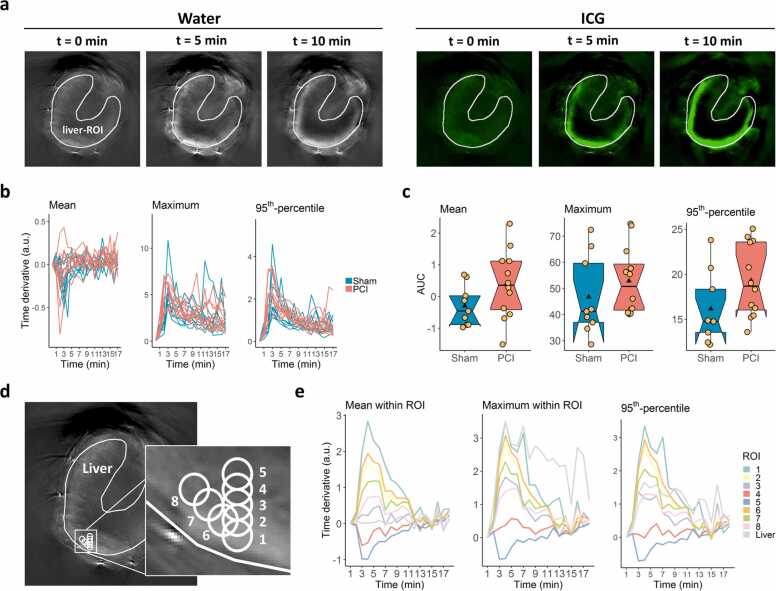
Fig. 5.
Tissue-oriented image analysis. (a) Liver-ROIs were drawn manually by an expert in the first time frame of the water channel of the MSOT image stacks (grey coloured images) and used to analyse signal intensity in the ICG channel (green coloured images). (b) The mean, maximum and 95th-percentile time derivative values within the liver-ROIs were extracted from the ICG channel. (c) AUC values of the time curves were calculated for each animal in the two treatment groups and are shown as boxplots for mean, maximum and 95th-percentile signal intensity values, with the black triangle denoting the average value and the yellow dots representing individual data points (mean: p = 0.09, g = 0.71; maximum: p = 0.22, g = 0.42; 95th-percentile: p = 0.049, g = 0.75; p-values were derived by two-tailed Wilcoxon rank sum test; g-values denote Hedges' g). (d) A ROI with a diameter of 10 pixels was drawn in the liver tissue region (ROI 1) and then shifted by 5-pixel steps in the y-direction (ROIs 2–5) or in the x-y-direction (ROIs 6–8) to obtain a set of ROIs within close proximity. (e) The mean, maximum and 95th-percentile signal intensity values from ROIs 1–8 and the liver-ROI were extracted from the pre-processed MSOT image and plotted for the whole time range.