Figure 1. ATP is crucial for both RNA-guided transposition of ShCAST, and filament assembly of AAA+ regulator TnsC.
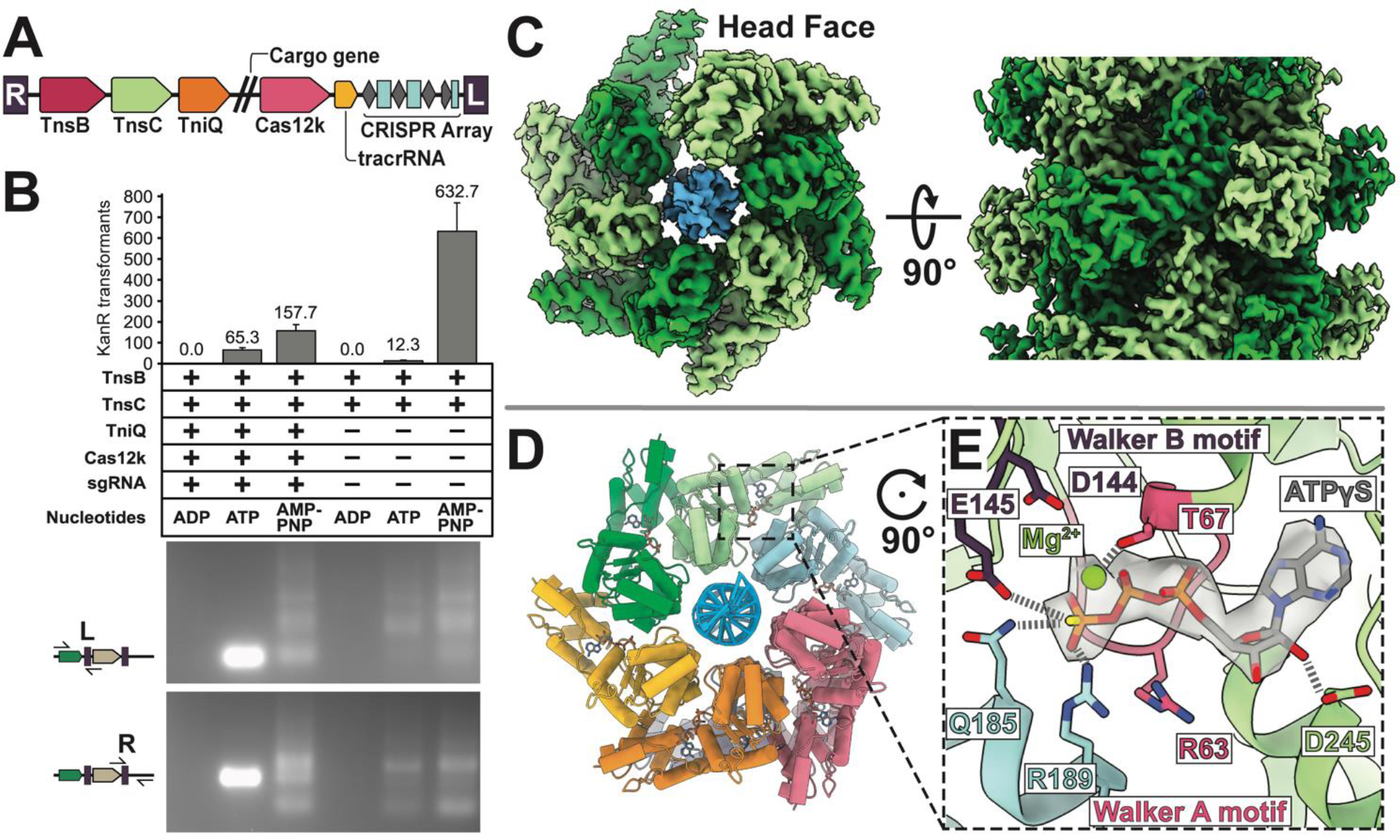
(A). The ShCAST transposon is defined by the right- (R) and left- (L) ends of the element, encoding: Cas12k RNA-binding effector, TniQ, TnsC, and TnsB, trans-activating crRNA (tracrRNA), and CRISPR array. Double slash represents the region where cargo genes are found in the transposon. (B) ATP-hydrolysis is required for targeted RNA-guided transposition. Colony counts from transformation of each deproteinated transposition reaction as a proxy for overall transposition activity are shown (one third of total reaction volume was used in transformation). Data indicate mean +/− standard deviation (n=3). Reaction mixes were additionally analyzed by PCR using a transposon end primer (L or R) along with primers flanking the target site as indicated in the schematic on the left. A high rate of on-target insertion results in a single intense band, while insertions distributed around the target plasmid results in a number of PCR products. A single representative image of n=3 replicates is shown. (C) High-resolution (3.2 Å) cryo-EM reconstruction of ATPγS-TnsC (light and dark green) forms a continuous helical filament encircling DNA (blue). The arrangement of layers (6 TnsC subunits) within the filament is referred to here as a ‘head-to-tail’ configuration. (D) Atomic model of TnsC helical filament consists of 6 subunits of TnsC arranged in a helical spiral. (E) The ATP-binding pocket follows canonical features of AAA+ proteins, with conserved Walker A (pink) and Walker B motif (purple) coordinating ATPγS-binding. The adjacent subunit (light blue) forms inter-subunit contacts that partially contribute to filament formation by forming interactions with the terminal phosphate.
