Abstract
The development of compounds with enhanced activity and selectivity by a conserved spatial orientation of the pharmacophore elements has a long history in medicinal chemistry. Rigidified compounds are an example of this concept. However, the intramolecular interactions were seldom used as a basis for conformational restraints. Here, we show the weak intramolecular interactions that contribute to the relatively well-conserved geometry of N1-arylsulfonyl indole derivatives. The structure analysis along with quantum mechanics calculations revealed a crucial impact of the sulfonyl group on the compound geometry. The weak intramolecular C–H⋯O interaction stabilizes the mutual "facing" orientation of two aromatic fragments. These findings extend the pharmacological interpretation of the sulfonyl group role from the double hydrogen bond acceptor to the conformational scaffold based on intramolecular forces. This feature has, to date, been omitted in in silico drug discovery. Our results should increase the awareness of researchers to consider the conformational preference when designing new compounds or improving computational methods.
The impact of weak intramolecular C–H⋯O interactions on the conformational stability of bis-arylsulfones is discussed, suggesting different role of sulfonyl group in the ligand – 5HT6 receptor interaction.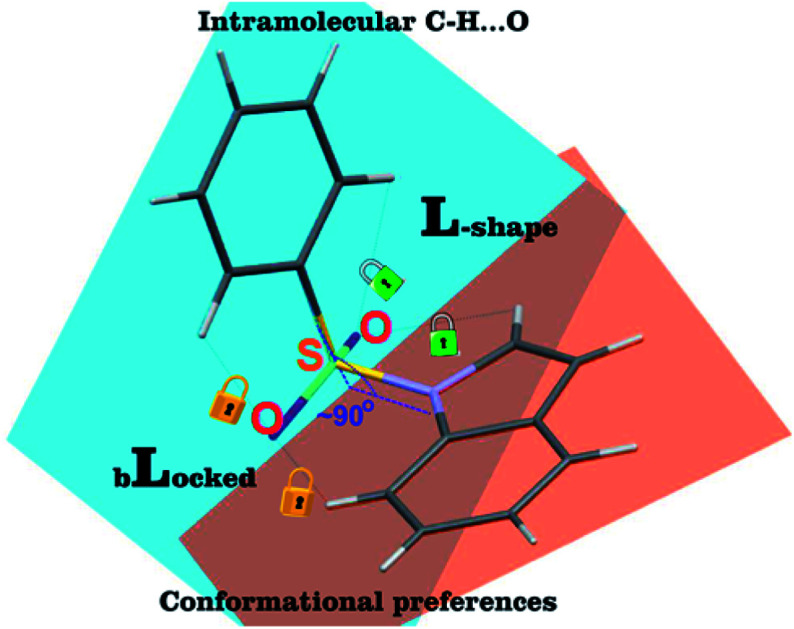
Introduction
The balance between rigidity and flexibility is a very important factor in drug discovery: more rigid molecules may exhibit better in vitro activity but worse pharmacokinetic properties than their flexible analogues. An effective way to overcome this issue is to introduce donors and acceptors of hydrogen bonds that allow the formation of intramolecular interactions. Such a strategy determines the shape of the molecule, which often leads to increased ligand-target affinity.1–6 This effect is generally caused by strong hydrogen bonds, but weak C–H⋯O interactions7 can also help to maintain the correct and active conformation.
Sulfones and sulfonamides are the most widely represented classes of the serotonin receptor subtype 6 (5-HT6R) ligands8–13 (87.8% (1817) of the total number (2069) of recognized 5-HT6R ligands with inhibition constants Ki (or IC50/2)14 ≤ 100 nM; ChEMBL database;15 ver. 23). Approximately 963 identified compounds are different sulfonamides and 873 ligands contain the sulfonyl group, which directly links two aromatic systems (one of them is usually a heterocyclic moiety, e.g., indole ring). It is worth mentioning that sulfones and sulfonamides are less abundant types of ligands for other targets of the serotonin receptor family, e.g., 5-HT1AR (3.7%), 5-HT2AR (7.5%) or 5-HT7R (25.4%) (ChEMBL database;15 ver. 23; presented values concern ligands with Ki (or IC50/2) ≤ 100 nM).
Among the ligands of 5-HT6R, the antagonists are especially valuable due to the procognitive and/or antiamnesic effects observed during several studies of age-related cognitive impairments, memory formations and memory deficits in schizophrenia, Parkinson disease or Alzheimer disease.12,16–21 Additionally, it has been reported, that the 5-HT6R ligands may be applied for the treatment of obesity.22 The commonly recognized pharmacophore of the 5-HT6R antagonists consists of four key fragments: basic nitrogen atom (PI), hydrophobic core (H), aromatic ring(s) (Ar) and dual acceptor of hydrogen bonds (HBA).23–28
In the pharmacophore model of the 5-HT6R antagonists, the sulfonyl fragment is usually considered as the strong hydrogen bond acceptor.23–25,27,28 Based on docking study, it is postulated that it interacts with N6.55 and S5.43 forming O–H⋯O and N–H⋯O bonds,23,27,28 respectively. However, in the statistical approach applied for the majority of protein–ligand complexes deposited within the PDB (Protein Data Bank), the sulfonyl moiety is not involved in the strong hydrogen bond formation and usually occupies the hydrophobic pocket of the protein biding site.29 Despite the electronegative character of this group, it usually forms weak hydrogen bonds with non-polar amino acid side chains and/or participates in several van der Waals interactions.
The sulfonyl linker appears to be essential in the structure–activity evaluation of bis-aromatic 5-HT6R antagonists. Attempts to replace this functional group with other fragments in most cases decreased the binding affinity to the receptor.4,8–11,30,31 Despite considering the HBA functionality of the sulfonyl group, its full impact on 5-HT6R ligand activity is still unclear.
In our recent study, we suggested that the key molecular characteristic of the bis-aromatic 5-HT6R antagonists is related to the precise mutual orientation of aromatic moieties.32 Similar observations were made for sulfonyl derivatives with the ability to form strong intramolecular hydrogen bonds, stabilizing the geometry of two aromatic fragments.4,9 To investigate the impact of the sulfonyl group on the conformational stability, selected N-arylsulfonyl indole/indazole derivatives, known 5-HT6R antagonists, were structurally studied. Bioisosteric replacement of the sulfonyl fragment was applied, including the methylene (tetrahedral conformation) and carbonyl (acceptor of the hydrogen bond) groups. Observations based on the crystal structure analysis supported by the Cambridge Crystallographic Database33 (CSD) statistics and theoretical calculations revealed an interesting geometric feature of the molecules in the most active sulfonyl derivatives, which was not observed in the bioisosteres. Although the impact of the sulfonyl group on conformational stability was already reported for different bioactive molecules (mostly sulfonamides),34–36 the theoretical explanation based on the intramolecular C–H⋯O interactions formation was not reported to date.
Materials and methods
X-ray structure determination and refinement
All sulfonyl derivatives for crystallization were synthesized 1a–4a10,37–39 along with bioisosters 1b–4b and 1c–4c (unpublished data). Crystals of indole/indazole derivative series (1–4) were obtained by slow evaporation of the solvent under ambient conditions. All obtained compounds were crystallized in the form of hydrochloride or oxalate salts. Only compound 1a was crystallized as a free base.
Intensities were collected using a SuperNova (Rigaku – Oxford Diffraction) four circle diffractometer with a mirror monochromator and a microfocus CuKα radiation source (λ = 1.5418 Å) for 1a and 1b and a MoKα radiation source (λ = 0.7107 Å) for all remaining compounds. Additionally, the diffractometer was equipped with the CryoJet HT cryostat system (Oxford Instruments), which allows low temperature experiments for crystals of compounds belonging to series 1, 3 and 4 (120–130 K). For derivatives of series 2, the experiments were performed at room temperature. The obtained data sets were processed with CrysAlisPro software.40 Absorption correction based on multiple scans was applied using spherical harmonics implemented in the SCALE3 ABSPACK scaling algorithm.40 The phase problem was solved by direct methods using SHELXS-2013/1.41 The parameters of the obtained models were refined by full-matrix least-squares on F2 using SHELXL-2014/6.41 All non-hydrogen atoms were refined anisotropically. H atoms attached to the protonated nitrogen atoms of the piperazine ring (structures 1b, 1c, series 3 and 4) or tertiary amine (series 2) and oxygen atoms (water, methanol or oxalate molecules) were located on the difference Fourier map.
WinGX42 software (ver. 2014.1) was used to prepare materials for publication. Figures presenting a single molecule extracted from the crystal environment as well as the superposition of molecules were generated with Mercury 3.7.43 To calculate the weighted least-squares planes through selected atoms, the PARST44 program was used. The detailed table showing crystallographic data and refinement descriptions are in Section 2 of the ESI.†
Theoretical calculations
The AIMAll package45 and NCIPLOT program46 were used to identify intramolecular interactions within the studied systems. To achieve this goal, we performed DFT calculations for isolated molecules with the GAUSSIAN09 47 package at the B3LYP/6-311++G(2d,2p) and B3LYP/6-311G** level. In both cases, the geometries were determined from X-ray diffraction data and kept frozen for calculations. Wave functions obtained with the 6-311++G(2d,2p) basis set were used to perform Bader's quantum theory of atoms (QTAIM)48 partitioning with the AIMAll package, and those obtained with the 6-311G** basis set were used to generate reduced electron density gradient (RDG) surfaces with the NCIPLOT program.
The NCI (non-covalent interaction) analysis is based on the reduced electron density gradient (RDG) defined as: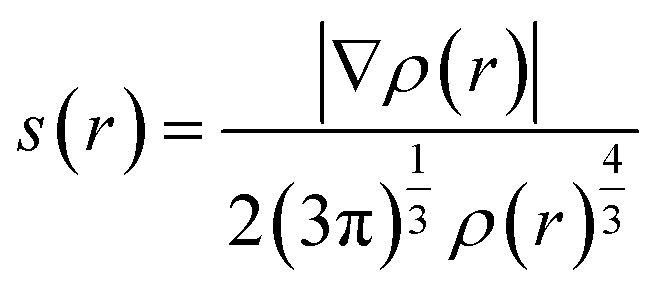 where ∇ρ(r) is a gradient of the electron density. This analysis enables visualization of regions involved in either attractive or repulsive interactions. If a non-covalent contact is present in the studied system on scatterplots of s(r) against ρ(r), characteristic spikes occur in low-gradient and low-density regions, which are absent when only covalent bonds are observed. Moreover, if the sign of the second eigenvalue (λ2) of the Hessian matrix of electron density is taken into account, it provides information concerning whether the identified non-covalent interaction is stabilizing (λ2 < 0) or destabilizing (λ2 > 0). Therefore, if a spike in the low-gradient, low-density area at negative λ2 is present, it indicates a stabilizing interaction such as a hydrogen bond, whereas when a smaller spike and slightly negative λ2 are observed, the interaction is weakly stabilizing, and finally when a spike is associated with a positive λ2, non-covalent interaction is absent. In many cases, if the peak representing NCI does not reach s(r) equal to 0, there is no associated critical point and therefore QTAIM is blind to this contact. The NCIPLOT plots were generated by evaluating the B3LYP 6-311G** density and reduced gradient on cuboid grids with a steo size 0.1 a.u.
where ∇ρ(r) is a gradient of the electron density. This analysis enables visualization of regions involved in either attractive or repulsive interactions. If a non-covalent contact is present in the studied system on scatterplots of s(r) against ρ(r), characteristic spikes occur in low-gradient and low-density regions, which are absent when only covalent bonds are observed. Moreover, if the sign of the second eigenvalue (λ2) of the Hessian matrix of electron density is taken into account, it provides information concerning whether the identified non-covalent interaction is stabilizing (λ2 < 0) or destabilizing (λ2 > 0). Therefore, if a spike in the low-gradient, low-density area at negative λ2 is present, it indicates a stabilizing interaction such as a hydrogen bond, whereas when a smaller spike and slightly negative λ2 are observed, the interaction is weakly stabilizing, and finally when a spike is associated with a positive λ2, non-covalent interaction is absent. In many cases, if the peak representing NCI does not reach s(r) equal to 0, there is no associated critical point and therefore QTAIM is blind to this contact. The NCIPLOT plots were generated by evaluating the B3LYP 6-311G** density and reduced gradient on cuboid grids with a steo size 0.1 a.u.
Cambridge Structural Database search
The statistical data for the conformational preferences of the N-arylsulfonyl substituted indole ring was derived by searching CSD Version 5.37 (November 2015)33 with the ConQuest 1.18 program.49 Two searches were performed only for organic compounds with the R factor set as ≤ 0.05. The searched fragments were defined as indole ring-substituted at N1 atom with a sulfonyl or methylene linker attached to the benzene ring with H atoms defined in both ortho positions (C12 and C16) (with respect to the sulfonyl or methylene fragments). The organometallic structures were excluded. The search resulted in 265 and 129 structures for sulfonyl and methylene linkers, respectively. From the group of structures with a methylene linker, four were excluded because they are macrocycles with a more rigid conformation. An additional search using the same parameterization was performed for structures with H atoms bound to carbon atoms C12–16 of the benzene ring and C2 and C8 of the indole moiety. Twenty seven and 37 structures containing sulfonyl and methylene linkers were found, respectively. From the chosen 27 structures of the sulfonyl derivatives, three were silica derivatives and were excluded due to additional effects influencing the geometry of the molecule.
Protein Data Bank search
The Protein Data Bank50 (www.rcsb.org) was searched for ligands with the option of chemical substructure similarity. Two searches were performed to identify bis-aromatic sulfonyl derivatives in complexes with different macromolecular targets. Ligands containing arylsulfonyl indole (3 ligands in 4 structures) or diphenylsulfone (22 ligands in 25 structures) substructures were selected for further investigation. From the first set, a ligand with ID JCB was excluded due to an unavailable PDB file, reducing the number to 2 ligands in 4 structures. In the second data set, ligand 6N0 (PDB ID: 5AKE) was excluded due to cyclization of the small molecule, which introduced rigidity affecting the investigated torsion angle.
Results and discussion
Binding affinity evaluation
Four known N-arylsulfonyl indole/indazole derivatives with confirmed antagonistic activity were selected and resynthesized (1a,102a,373a38 and 4a,39 see Table 1) along with their bioisosteres (replacement of the sulfonyl fragment by carbonyl group 1b, 2b, 3b and 4b and by methylene linker 1c, 2c, 3c and 4c). The binding affinities towards 5-HT6R were high for all obtained sulfonyl derivatives, with Ki ranging from 1–11 nM (Table 1). Substitution of the sulfonyl group with the carbonyl linker caused, in most cases, a significant drop in affinity, while introduction of the methylene linker led to a less significant decrease in the Ki values. The binding affinities towards 5-HT1AR, 5-HT7R and D2R (dopamine receptor) were also determined, confirming the selective profile of the sulfonyl derivatives (ESI, Section S1†).
Molecular structures and binding affinities of the tested compounds towards 5-HT6R.
| General structure | No. | L | 5-HT6R Ki [nM]a |
|---|---|---|---|
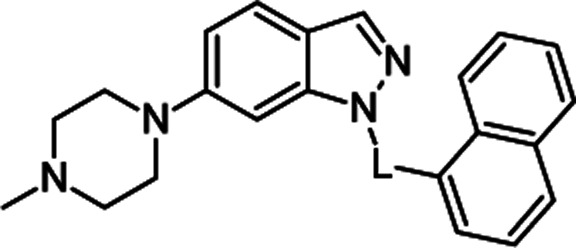
|
1a | –SO2– | 1 ± 0.2 |
| 1b | –CO– | 1280 ± 231 | |
| 1c | –CH2– | 51 ± 7 | |
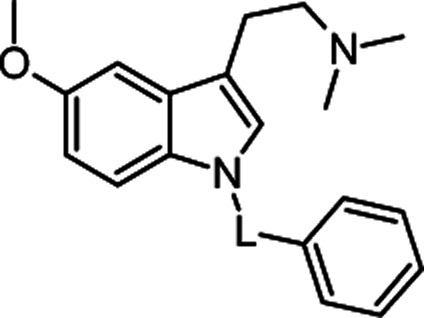
|
2a | –SO2– | 11 ± 2 |
| 2b | –CO– | 44 ± 8 | |
| 2c | –CH2– | 23 ± 5 | |
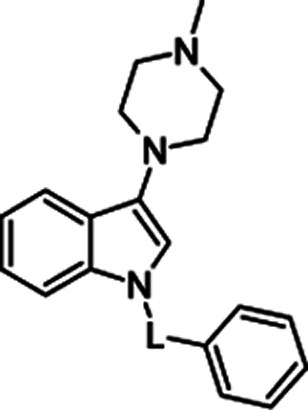
|
3a | –SO2– | 4 ± 1 |
| 3b | –CO– | 187 ± 22 | |
| 3c | –CH2– | 18 ± 3 | |
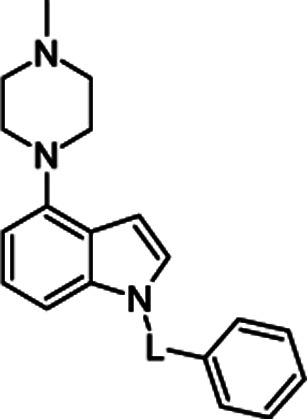
|
4a | –SO2– | 1 ± 0.3 |
| 4b | –CO– | 2204 ± 189 | |
| 4c | –CH2– | 24 ± 5 |
To test the selectivity profile of presented ligands towards 5-HT6R, the binding affinities towards 5-HT1AR, 5-HT7R and D2R were additionally determined for selected compounds. For experimental details and measured values, see Section S1 in the ESI.
Crystal structure analysis
To investigate the observed structure–activity relationship (SAR), the X-ray crystal structure analysis was applied to demonstrate structural differences and similarities among all presented compounds. Molecule 3b was excluded from the study due to unsuccessful crystallization trials.
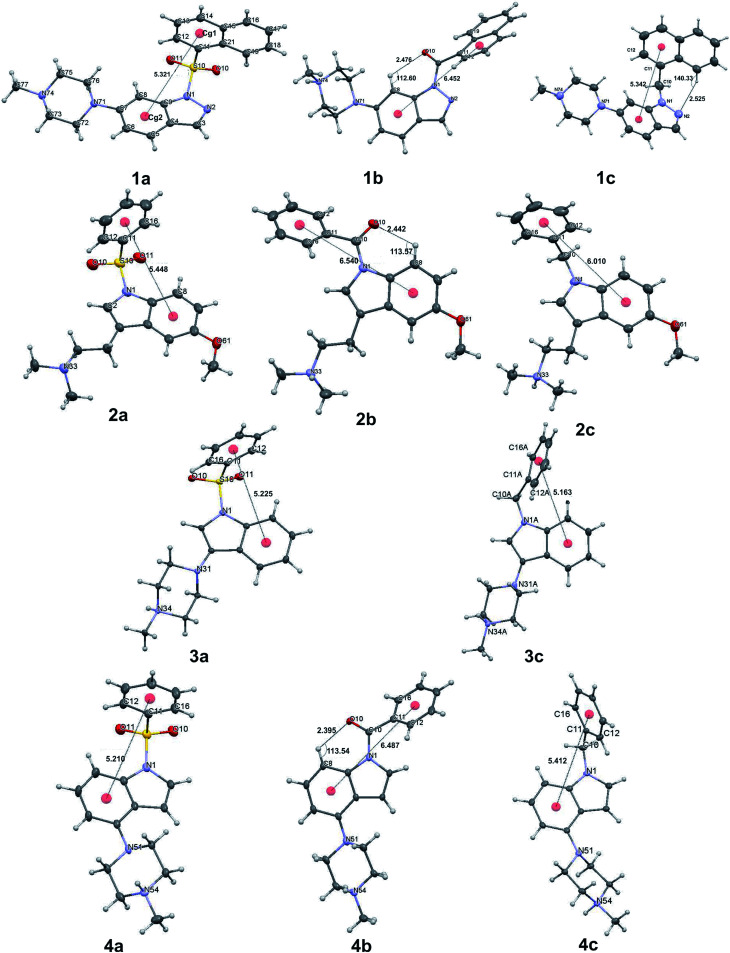
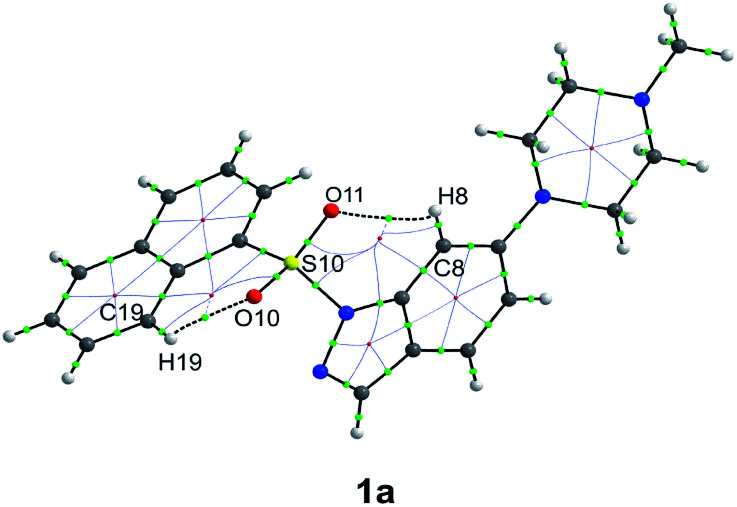
All but one (1a) of the presented structures crystallize in the salt forms (oxalates or chlorides), with positive charge (corresponding to the PI pharmacophore feature) located on the external, most basic nitrogen atom of the piperazine or non-rigid tertiary amine group (detailed description of all crystal structures is presented in Section S2 of the ESI†). The conformations of a single molecule extracted from the crystal structure are shown in Fig. 1.
Fig. 1. Conformation of the single molecule extracted from the crystal environment (for 3a, 4a and 4b), only one molecule of the asymmetric unit is shown.
The asymmetric unit of 3c consists of two molecules in distinct conformations, whereas in the case of the crystal structure of 4a and 4b there are two and four similar conformers, respectively (ESI Sections 2.3 and 2.4†). Thus, only in the case of structure 3c, each molecule was treated as a separate conformer. To demonstrate the conformational divergence of the presented bioisostere sets, a superposition of the indole/indazole ring was performed (ESI, Sections 2.2–2.5†). The conformation of compounds with a carbonyl linker is most distinct and is stabilized by the intramolecular C8–H8⋯O10 hydrogen bond (Fig. 1). As a consequence, the spatial orientation of the two aromatic fragments in carbonyl derivatives differs significantly with respect to derivatives with sulfonyl and methylene linkers. Such divergent conformation may explain the observed decrease in the binding affinity and suggests that the mutual orientation of both aromatic moieties may be a very important factor for bioactivity.
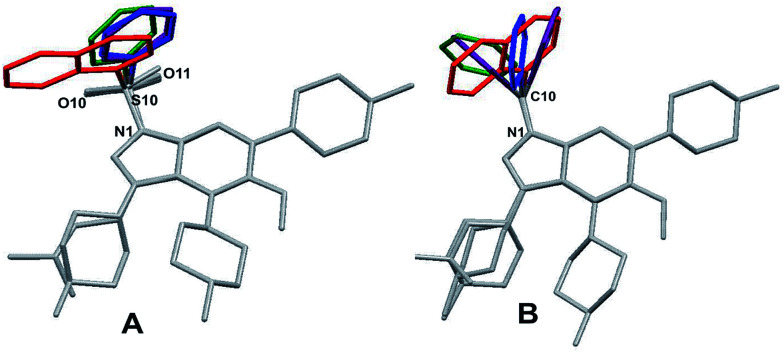
In contrast, the conformation of molecules with sulfonyl and methylene linkers are more similar, allowing only different rotational positioning of the phenyl ring against the indole/indazole fragment. This observation may explain the small drop in binding affinity for bioisosteres with the methylene linker. Superposition of the molecules extracted from the crystal structures with the same linker type (1a, 2a, 3a, 4a for sulfonyl and 1c, 2c, 3c, 4c for methylene) revealed an additional structural feature and conformational preferences (Fig. 2), which may be a critical factor, with a high influence on the 5-HT6R binding affinity.
Fig. 2. Conformations observed in the crystal structure, superposed with respect to the indole/indazole ring, showing the preferential orientation of the aromatic moiety for sulfonyl (A) and its random distribution for methylene (B) derivatives (for clarity, only the aromatic fragments were distinguished with the same color for corresponding pairs of bioisosteres: 1a and 1c (red), 2a and 2c (green), 3a, 3c and 3c′ (purple), 4a and 4c (blue)).
In the arylsulfonyl derivatives presented herein, both aromatic rings of the molecule had a relatively similar L-shape orientation, allowing two π-electron systems to remain in proximity (Fig. 1, structures 1a–4a, Fig. 2). The phenyl/naphthyl ring is slightly twisted, facing the six-membered ring of the indole/indazole moiety. Despite the tetrahedral spatial arrangement of the methylene linker, similar structural preferences of the orientation of aromatic moieties were not observed (Fig. 1, structures 1c, 2c, 3c and 4c, Fig. 2).
A detailed analysis of distances and angles revealed numerical factors that are consistent for most active 5-HT6R ligands, e.g., the distance between centroids Cg1 assigned for the six-membered ring of the indole/indazole and Cg2 defined for the six-membered phenyl/naphthyl ring directly bound to the sulfonyl linker, which stays in the range from 5.1–5.5 Å (Fig. 1). This parameter is in agreement with our previously published data for non-sulfonyl derivatives.32
Another parameter is the angle between the mean planes44,51 defined by atoms of both aromatic fragments (PLN1 and PLN2 for indole/indazole and phenyl/naphthyl rings, respectively), which are in an approximately perpendicular orientation (angle ranging from 80–90°). The final parameter is the C12–C11–S10(or C10)–N1 torsion angle, which for most active sulfonyl derivatives stays in the range from 70–100° and seem to be the most prominent factor determining the L-shape orientation of both aromatic fragments (for a comparison of geometric parameters see table in the ESI, Section S2.6†).
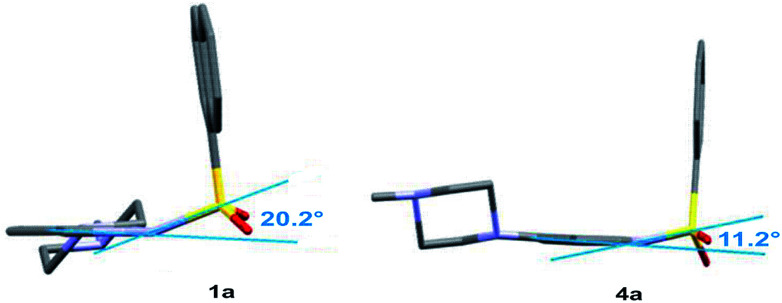
The structural feature that additionally supports the more bent conformation of the sulfonyl derivatives and also distinguishes them from other bioisosteres is the pyramidality of nitrogen atom N1 of the indole/indazole ring. As previously reported,52,53 the sulfonyl group does not exhibit a strong withdrawing effect, leading to an angular position of the sulfur atom with respect to the indole/indazole plane (PLN1) (the angular deviations of the S10 or C10 atom of PLN1 as well as the external angles between the S10(C10)–N1 bond and PLN1 are presented in the ESI, Section S2.6†) the highest angular deviation of the linking sulfur atom is observed for the indazole derivative 1a (20.2°, Fig. 3). According to this observation, the sulfonyl linker allows a more aromatic characteristic of the indole/indazole system in comparison to the obtained bioisosteres. For all presented derivatives with a methylene linker, the C10 atom is placed more planar with respect to the indole/indazole plane.
Fig. 3. The angular deviation and pyramidalization of the indazole/indole N1 atom observed in the crystal structure of compounds 1a and 4a.
Cambridge Structural Database analysis
To test if the observed conformational preferences are characteristic for similar sulfonyl derivatives, the Cambridge Crystallographic Database (CSD) statistical search was performed for all deposited N-substituted arylsulfonyl and benzyl indole derivatives (with unsubstituted ortho positions C12 and C16 of the phenyl ring). The search resulted in 265 and 125 structures with sulfonyl and methylene linkers, respectively, which were subsequently evaluated to search for preferences in the mutual orientation of the aromatic moieties. The tested geometric parameters were the Cg1⋯Cg2 distance, PLN1-PLN2 angle and the torsion angle corresponding to C12–C11–S10(or –C10)–N1. For the purposes of this search, a variety of substituents attached to the indole moiety were included in the statistics. The type of substituent, its position in the indole ring, possible steric hindrance and the specific interactions in the crystal lattice clearly have a great impact on the presented statistical results. The mean distances Cg1⋯Cg2 in the CSD sets is 5.3 Å and 5.5 Å for structures with the sulfonyl and methylene linker, respectively. Those values are in agreement with the compounds presented in this paper. The mean values of the PLN1-PLN2 angle, at approximately 81° and 82° for arylsulfonyl and benzyl derivatives deposited within the CSD, are in the predicted range for the best 5-HT6R ligands. It is worth noting that the statistical distribution of both above-mentioned parameters in bis-aromatic structures with sulfonyl and methylene linkers are similar (ESI, Section S4.1†).
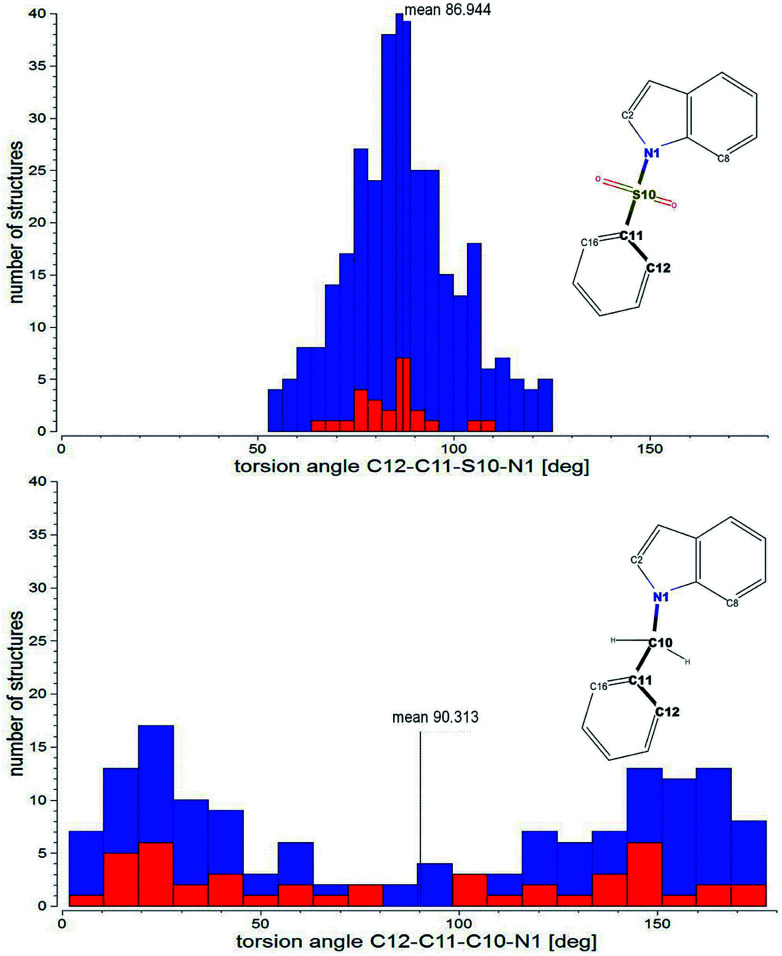
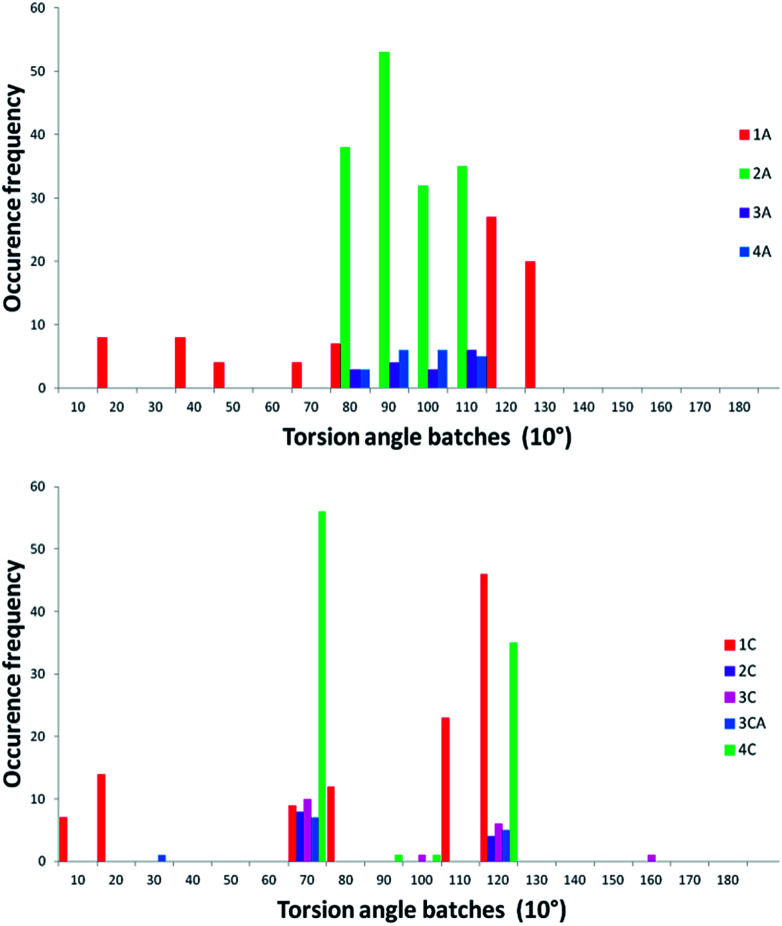
The main parameter describing the mutual orientation of the aromatic rings, but also differentiating both types of tested compounds, is the torsion angle corresponding to C12–C11–S10(or –C10)–N1. The distribution of this torsion angle among all investigated structures found in the CSD demonstrates a strong preference in the aromatic fragments orientation for sulfonyl derivatives and a random arrangement for compounds with the methylene linker (Fig. 4). Moreover, the dominant range of the torsion angle for sulfonyl derivatives seems to make up only a small proportion of the compounds with a methylene linker.
Fig. 4. Distribution of the corresponding torsion angle: C12–C11–S10(or –C10)–N1 observed in the crystal structures of indole derivatives deposited within CSD with a sulfonyl (upper histogram – 265 structures) or methylene (lower histogram – 125 structures) linker (search was performed for molecules with hydrogen atoms defined at C12 and C16 of the benzene ring). Red bars show the distribution of the torsion angle in the C2 and C8 unsubstituted indole moiety (24 and 37 structures for sulfonyl and methylene derivatives, respectively).
For structures from the CSD, the indole nitrogen pyramidalization effect is much more profound in the presence of the sulfonyl than the methylene linker. The angular deviation of the S10 atom from the mean plane of the indole ring (PLN1) ranges from 0–59°, (mean value = 15°), whereas for the methylene linker, the deviation of C10 varies from 0° up to 22° (mean value = 6°). In contrast, both mentioned atoms are nearly in the plane of the benzene ring (PLN2) (mean value = 1.7° and 1.9° for the sulfur (S10) and carbon (C10) atom, respectively).
According to the structural observation, we postulate that the main forces allowing the two aromatic fragments to remain in the preferential orientation are weak intramolecular hydrogen bonds. These hydrogen bonds are additionally supported by close contacts of the C–H⋯O type (Table 2). Moreover, this intramolecular interaction may be characteristic of different types of bis-aromatic systems that are linked directly to sulfonyl group (for example, CSD searches of diphenyl sulfone and diphenylmethane derivatives have been performed, confirming the conformational preferences for sulfonyl linker – ESI, Section S4.2†).
Geometric parameters of selected intramolecular hydrogen bondsa and close contacts defined for arylsulfonyl derivative molecules.
| No. | D–H⋯A | d(D⋯A) [Å] | d(H⋯A) [Å] | D–H⋯A [°] |
|---|---|---|---|---|
| 1a | C8–H8⋯O11 | 3.004(4) | 2.440(2) | 117.9(2) |
| C12–H12⋯O11 | 2.861(3) | 2.457(2) | 105.5(2) | |
| C19–H19⋯O10 | 2.914(4) | 2.235(2) | 127.6(2) | |
| 2a | C8–H8⋯O11 | 3.115(2) | 2.577(1) | 117.3(1) |
| C12–H12⋯O11 | 2.891(2) | 2.514(1) | 104.5(1) | |
| 3a | C8–H8⋯O11 | 3.096(2) | 2.545(1) | 117.2(1) |
| C12–H12⋯O11 | 2.902(2) | 2.522(1) | 104.0(1) | |
| 4a | C8–H8⋯O11 | 3.102(3) | 2.548(2) | 117.4(2) |
| C12–H12⋯O11 | 2.958(3) | 2.609(1) | 102.1(1) | |
| C16–H16⋯O10 | 2.945(3) | 2.600(2) | 101.8(1) |
Contacts with the geometrical parameters of the hydrogen bond are indicated in bold.
QTAIM and NCI approach to intramolecular H-bond evaluation
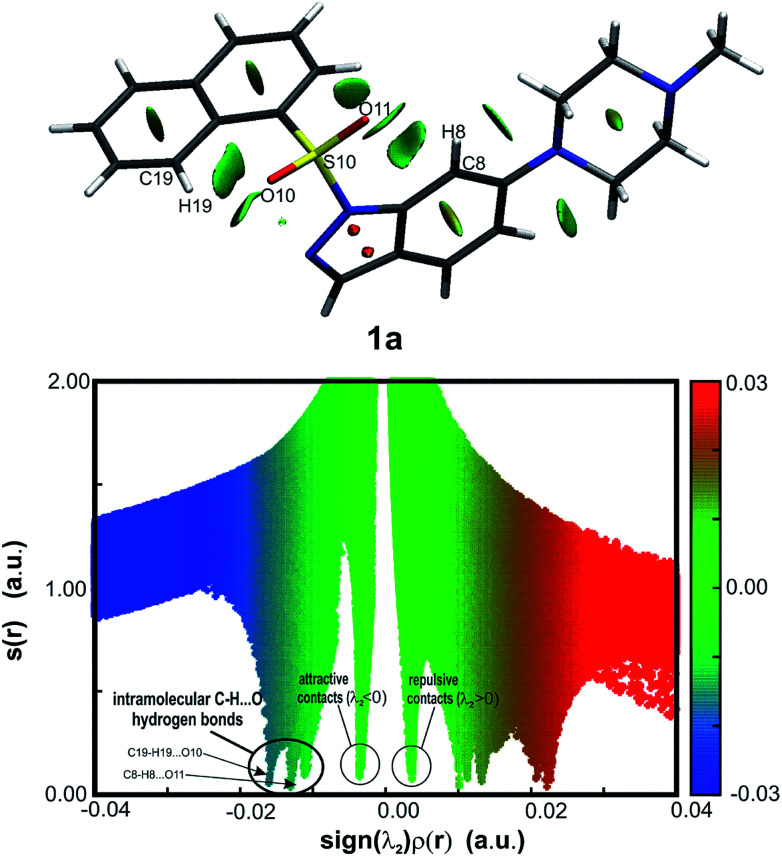
The routinely used approach to assess hydrogen bond formation is based on the atoms in molecule theory (AIM).48 To reveal the impact of intramolecular forces on the molecular conformation of the studied systems, Bader's quantum theory of atoms (QTAIM)48 was applied (AIMAll package45). The topological analysis of the electron density ρ(r) of sulfonyl derivatives reveals Bond Critical Points (BCPs) accompanied by Ring Critical Points (RCPs) (as an example, structure 1a is shown in Fig. 5, for 2a–4a see ESI, Fig. 5.1 in Section S5;† BCPs and RCPs are shown as green and red dots, respectively), indicating the weak intramolecular interaction C8–H8⋯O11. Due to a more expanded naphthalene moiety in 1a, an additional weak hydrogen bond C19–H19⋯O10 is recognized. The calculated hydrogen bond energies54 vary from −1.9 to −2.5 kcal mol−1 for the C8–H8⋯O11 interaction (moderate weak hydrogen bond), whereas for C19–H19⋯O10 the energy is −3.5 kcal mol−1 (strong C–H⋯O interaction).
Fig. 5. Molecular graph of isolated molecule 1a (as an example) showing the bond paths and Bond Critical Points (BCP) in the studied system. The small green and red spheres indicate the (3, −1) BCP and (3, +1) RCP (Ring Critical Point) in ρ(r), respectively. Dashed bonds show weak intramolecular interactions of the C–H⋯O type.
As the QTAIM analysis is not sufficient to detect all expected weak non-covalent interactions, especially intramolecular hydrogen bonds,55 the NCI (non-covalent interaction) approach56,57 was additionally applied (NCIPLOT46). The isosurfaces of the reduced density gradient s(r) were examined, and corresponding plots were generated for the presented sulfonyl derivatives (as an example, structure 1a is shown in Fig. 6, for 2a–4a see ESI, Fig. 5.2 and 5.3, Section S5†). All sulfonyl derivatives exhibit evident weak attractive non-covalent interactions between indole or indazole ring atoms and O11, which are absent for compounds with the methylene linker (see ESI, Fig. 5.4 and 5.5, Section S5†). The exception is compound 1c, for which the C19–H19⋯N2 intramolecular hydrogen bond is observed (EHB = −1.6 kcal mol−1) as a consequence of the acceptor properties of the indazole nitrogen atom N2 (ESI, Fig. 5.6 and 5.7, Table 5.2, Section S5†). To identify spike positions in the s(r) vs. sign(λ2)ρ(r) plot, the unsubstituted 1-phenylsulfonyl-indole structure (CCDC ID: DUPTEN)52 was analyzed using the NCI approach (ESI, Fig. 5.8 and 5.9, Section 5†).
Fig. 6. Gradient isosurfaces representing intramolecular interactions (top) and corresponding plots of the reduced density gradient (RDG) vs. the electron density multiplied by the sign of the second Hessian eigenvalue (bottom) for the isolated molecule 1a (as an example). The shape of the domains is correlated with the strength of the interaction. Broad green/blue disk-shaped domains indicate weak attractive (λ2 < 0) and green repulsive (λ2 > 0) interactions (represented as blue/green and green spikes in the plot below). Delocalized electrons of aromatic groups are represented by egg-shaped domains. Gradient surfaces correspond to s = 0.1 a.u. The color scale in the plot corresponds to −0.04 < ρ < 0.04 a.u.
Although QTAIM analysis of the sulfonyl derivatives reveals C19–H19⋯O10 hydrogen bond formation only in the case of compound 1a, the isosurface analysis in Fig. 6 suggests that weaker interactions, especially in the proximity of the O11 atom, are also present in other molecules. The observed attractive and repulsive contacts are in equilibrium, allowing two aromatic moieties of the molecule to remain in the defined mutual orientation.
Protein Data Bank (PDB) analysis
The conformational preferences of bis-aromatic sulfonyl derivatives in complexes with different macromolecular targets were also investigated for structures deposited in the PDB50 (www.rcsb.org). Ligands containing arylsulfonyl indole (2 ligands in 4 structures) or diphenylsulfone (21 ligands in 24 structures) moieties were selected for further investigation (ESI, Section 6†). Despite different substituent types and possible steric hindrance in the selected ligands, the majority of structures exhibit a nearly perpendicular orientation of the aromatic moieties (torsion angles corresponding to C12–C11–S10–N1 range from approx. 65–90°). Only 3 ligands have significantly different geometries of the studied molecular fragment (ligand PDB ID/torsion angle: WDT58/52°; 2QJ59/45° and D3F60/22°). Detailed inspection of the electron density and difference Fourier maps around the organic molecule in the protein–ligand complex revealed that in the case of WDT, one of the aromatic rings is not precisely fitted in the electron density and for 2QJ an alternative position of the m-bromobenzene is possible, which was deduced based on positive peak on the difference Fourier map, indicating a rotational conformer. The geometry of the D3F ligand is unusual and uncertain because this molecule does not fit in the electron density, exhibiting larger than expected maxima and minima in the difference Fourier map (undetermined position). The density maps inspection for these three abovementioned structures with unusual ligands conformation suggests that the molecular geometry of these bis-aromatic sulfonyl compounds is highly uncertain.
The binding mode inspection of all arylsulfonyl indole and diphenylsulfone derivative PDB complexes (in total 28 crystal structures) revealed only one structure (PDB ID: 3OQ1 (ref. 61)) in which sulfonyl group serves as a strong hydrogen bond acceptor (with A172 main chain amine group as a donor). In all remaining structures, the sulfonyl group occupies hydrophobic pockets, what stays in agreement with Bissantz, et al.29
Conformer prediction
In modern in silico drug discovery, docking studies that rely on a conformational search for the ligand are routinely used.62–64 In this case, energy minimization of the system is required, but in our experience, many force fields neglect the conformational preferences based on weak intramolecular interactions and often lead to unrealistic ligand geometries.
For compounds 1a–4a and 1c–4c, the conformational search was calculated using a MacroModel module from Schrödinger Suite 2016-4.65 For predicted conformers, the torsion angles C12–C11–S10(or –C10)–N1 were measured and presented in histogram form (Fig. 7). For compounds 2a, 3a and 4a, the distribution of the torsion angle is consistent with the experimentally derived data (Fig. 4). Surprisingly, conformation of indazole derivative 1a exhibits a more random distribution of the selected torsion angle in predicted conformers, with no representatives in the experimentally and theoretically proven favorable range from 90°–110°.
Fig. 7. Distribution of the C12–C11–S10(or –C10)–N1 torsion angle in predicted/calculated conformers of compounds 1a–4a (upper) and 1c–4c (lower). Frequencies are estimated for 10° step batches.
Interestingly, the calculated conformations of the indole/indazole derivatives with the methylene linker are not randomly distributed as in the CSD histogram (Fig. 4), and are mostly represented by geometries with the C12–C11–C10–N1 torsion angle assembled in two batches at approximately 70° and 120° (corresponding values).
Conclusions
A statistical approach based on X-ray structure analysis, supported by quantum mechanics calculations was applied to explore the conformational preferences observed in N1-arylsulfonyl indole/indazole derivatives. Structural investigation of the 5-HT6R antagonists reveal a conformational preference underlying the high potency and selectivity of this classical 5-HT6R antagonist core. According to our findings, ligands with sulfonyl linkers exhibit the conformational preference in the mutual aromatic ring orientation, which may explain their stronger binding affinity and higher selectivity for the receptor, whereas the N1-benzyl indole derivatives are more flexible, which is less favored for selective 5-HT6R antagonists. The observed sofa-like conformation of the sulfonyl derivatives does not prevent the plausible hydrogen bond formation with N6.55 and S5.43. [Binding modes/ligand's geometries of 3a and 3c (as an example) in the 5HT6R binding site are shown in ESI, Section S1.3†] Moreover, such interactions can have an additional stabilizing effect on the ligand–protein complex formation. The methylene linker, equally, can interact with both abovementioned amino acids. However, in this case, it would be a donor of weak hydrogen bonds (most probably C–H⋯O type). That would explain lower, but still significant affinity of the N1-benzyl indole derivatives. Thus, the L-shape orientation of the two aromatic fragments may be a strong 5-HT6R ligands pharmacophore feature, irrespective of the lack or presence of the sulfonyl group, as reported in other studies.32
However, expanding the analysis beyond the 5-HT6R ligands justifies the claim that the conformational stability can be extrapolated to other bis-aromatic compounds linked to a sulfonyl group, as confirmed in a more statistically random set of derivatives deposited in the CSD as well as in conformations of corresponding ligands observed in the binding sites of protein crystal structures deposited in the PDB. The observed stabilization effect is based on the weak intramolecular C–H⋯O hydrogen bond formation. Its strength and stability is dependent on the position and withdrawing/donating effects of substituents attached to the aromatic fragment. Interestingly, the naphthyl ring forms the unexpectedly strong hydrogen bond C19–H19⋯O10 (EHB = −3.451 kcal mol−1), additionally stabilizing the L-shape conformation. Engagement of the oxygen lone pairs in the intramolecular interactions can explain the observed reduced electronegative character of the sulfonyl group. This observation supports the finding that in the majority of protein–ligand complexes in the PDB, this functional group is less involved in strong hydrogen bonding.29
In contrast, modern in silico drug discovery relies on a conformational search of the ligand (especially in the case of docking studies) that requires energy minimization of the system.62–64 However, in our experience, many force fields neglect weak intramolecular interactions and conformational preferences of the ligand,66 often leading to unrealistic predicted geometries of the ligand in the binding site. Bearing this information in mind, this research should raise the awareness of researchers and potentially impact the docking algorithms, which tend to assume that the sulfonyl group is a strong hydrogen bond acceptor. Additionally, given the imperfect conformational search tools, computational experiments (especially docking) with bis-aromatic compounds linked to sulfonyl should be meticulously inspected, and pose filtering with geometric constraints should probably be undertaken.
Conformational preferences identified among sulfonyl-linked bis-aromatic compounds may contribute to the refinement of the pharmacophore models, especially for 5-HT6R, which in consequence may lead to designing new drugs with better pharmacological profile. Also for other putative drugs, the weak intramolecular interactions (including C–H⋯O) along with all well defined conformational effects can be thoughtfully investigated before active conformer prediction to increase the success rate. Additionally, understanding of the intramolecular forces can be applied in designing of the new crystal forms and predicting polymorphism effects (crystal engineering).7
Conflicts of interest
Authors declare no conflicts of interest.
Supplementary Material
Acknowledgments
The study was partially supported by the grant OPUS 2014/13/B/NZ7/02210 from the Polish National Science Centre. The research was carried out with the equipment purchased thanks to the financial support of the European Regional Development Fund in the framework of the Polish Innovation Economy Operational Program (contract no. POIG.02.01.00-12-023/08). The theoretical calculations were supported in part by PL-Grid Infrastructure.
Electronic supplementary information (ESI) available: The Supporting Information file consists of paragraphs: S1 Binding affinity determination; S2. X-ray structure analysis; S3. Mass spectroscopy data of the dissolved crystal 3a; S4. Cambridge Crystallographic Database statistical analysis; S5. Theoretical calculations (additional QTAIM & NCI); S6. Protein Data Bank analysis. CCDC 1550187 (1a), CCDC 1550183 (1b), CCDC 1550184 (1c), CCDC 1550185 (2a), CCDC 1550186 (2b), CCDC 1550191 (2c), CCDC 1550188 (3a), CCDC 1550189 (3c), CCDC 1550190 (4a), CCDC 1550192 (4b), CCDC 1550193 (4c). Copies of the data can be obtained, free of charge, on application to CCDC, 12 Union Road, Cambridge CB2 1EZ, UK, (fax: +44-(0)1223-336033 or e-mail: deposit@ccdc.cam.ac.uk). For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c8ra03107j
References
- Kuhn B. Mohr P. Stahl M. J. Med. Chem. 2010;53:2601–2611. doi: 10.1021/jm100087s. [DOI] [PubMed] [Google Scholar]
- Bohm H. Klebe G. Böhm H. J. Angew. Chem., Int. Ed. Engl. 1996;35:2588–2614. doi: 10.1002/anie.199625881. [DOI] [Google Scholar]
- Foloppe N. Chen I. J. Curr. Med. Chem. 2009;16:3381–3413. doi: 10.2174/092986709789057680. [DOI] [PubMed] [Google Scholar]
- Ivachtchenko A. V. Golovina E. S. Kadieva M. G. Kysil V. M. Mitkin O. D. Tkachenko S. E. Okun I. Bioorg. Med. Chem. 2011;19:1482–1491. doi: 10.1016/j.bmc.2010.12.055. [DOI] [PubMed] [Google Scholar]
- Schärfer C. Schulz-Gasch T. Ehrlich H. C. Guba W. Rarey M. Stahl M. J. Med. Chem. 2013;56:2016–2028. doi: 10.1021/jm3016816. [DOI] [PubMed] [Google Scholar]
- Tao Z. F. Wang L. Stewart K. D. Chen Z. Gu W. Bui M. H. Merta P. Zhang H. Kovar P. Johnson E. Park C. Judge R. Rosenberg S. Sowin T. Lin N.-H. J. Med. Chem. 2007;50:1514–1527. doi: 10.1021/jm061247v. [DOI] [PubMed] [Google Scholar]
- Desiraju G. R. and Steiner T., The Weak Hydrogen Bond in Structural Chemistry and Biology, IUCr, Oxford University Press, Oxford, UK, 1999 [Google Scholar]
- Ivachtchenko A. V. Golovina E. S. Kadieva M. G. Koryakova A. G. Mitkin O. D. Tkachenko S. E. Kysil V. M. Okun I. M. Eur. J. Med. Chem. 2011;46:1189–1197. doi: 10.1016/j.ejmech.2011.01.038. [DOI] [PubMed] [Google Scholar]
- Ivachtchenko A. V. Golovina E. S. Kadieva M. G. Kysil V. M. Mitkin O. D. Tkachenko S. E. Okun I. M. J. Med. Chem. 2011;54:8161–8173. doi: 10.1021/jm201079g. [DOI] [PubMed] [Google Scholar]
- Liu K. G. Lo J. R. Comery T. A. Zhang G. M. Zhang J. Y. Kowal D. M. Smith D. L. Di L. Kerns E. H. Schechter L. E. Robichaud A. J. Bioorg. Med. Chem. Lett. 2009;19:2413–2415. doi: 10.1016/j.bmcl.2009.03.071. [DOI] [PubMed] [Google Scholar]
- Liu K. G. Lo J. R. Comery T. A. Zhang G. M. Zhang J. Y. Kowal D. M. Smith D. L. Di L. Kerns E. H. Schechter L. E. Robichaud A. J. Bioorg. Med. Chem. Lett. 2009;19:3214–3216. doi: 10.1016/j.bmcl.2009.04.108. [DOI] [PubMed] [Google Scholar]
- Liu K. G. Robichaud A. J. Drug Dev. Res. 2009;70:145–168. doi: 10.1002/ddr.20293. [DOI] [Google Scholar]
- Liu K. G. Robichaud A. J. Greenfield A. A. Lo J. R. Grosanu C. Mattes J. F. Cai Y. Zhang G. M. Zhang J. Y. Kowal D. M. Smith D. L. Di L. Kerns E. H. Schechter L. E. Comery T. A. Bioorg. Med. Chem. 2011;19:650–662. doi: 10.1016/j.bmc.2010.10.033. [DOI] [PubMed] [Google Scholar]
- Kalliokoski T. Kramer C. Vulpetti A. Gedeck P. PLoS One. 2013;8:e61007. doi: 10.1371/journal.pone.0061007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bento A. P. Gaulton A. Hersey A. Bellis L. J. Chambers J. Davies M. Krüger F. A. Light Y. Mak L. McGlinchey S. Nowotka M. Papadatos G. Santos R. Overington J. P. Nucleic Acids Res. 2014;42:D1083–D1090. doi: 10.1093/nar/gkt1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M. V. Marsden C. A. Fone K. C. F. Trends Pharmacol. Sci. 2008;29:482–492. doi: 10.1016/j.tips.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Huerta-Rivas A. Pérez-García G. González-Espinosa C. Meneses A. Neurobiol. Learn. Mem. 2010;93:99–110. doi: 10.1016/j.nlm.2009.08.009. [DOI] [PubMed] [Google Scholar]
- Meneses A. Pérez-García G. Ponce-Lopez T. Castillo C. Int. Rev. Neurobiol. 2011;96:27–47. doi: 10.1016/B978-0-12-385902-0.00002-4. [DOI] [PubMed] [Google Scholar]
- de Bruin N. M. W. J. Kruse C. G. Curr. Pharm. Des. 2015;21:3739–3759. doi: 10.2174/1381612821666150605112105. [DOI] [PubMed] [Google Scholar]
- Claeysen S. Bockaert J. Giannoni P. ACS Chem. Neurosci. 2015;6:940–943. doi: 10.1021/acschemneuro.5b00135. [DOI] [PubMed] [Google Scholar]
- Ramirez M. J. Lai M. K. P. Tordera R. M. Francis P. T. Drugs. 2014;74:729–736. doi: 10.1007/s40265-014-0217-5. [DOI] [PubMed] [Google Scholar]
- Woolley M. Bentley J. Sleight A. Marsden C. Fone K. C. Neuropharmacology. 2001;41:210–219. doi: 10.1016/S0028-3908(01)00056-9. [DOI] [PubMed] [Google Scholar]
- Lopez-Rodriguez M. L. Benhamu B. de la Fuente T. Sanz A. Pardo L. Campillo M. J. Med. Chem. 2005;48:4216–4219. doi: 10.1021/jm050247c. [DOI] [PubMed] [Google Scholar]
- Kim H. J. Doddareddy M. R. Choo H. Cho Y. S. No K. T. Park W. K. Pae A. N. J. Chem. Inf. Model. 2008;48:197–206. doi: 10.1021/ci700160t. [DOI] [PubMed] [Google Scholar]
- V Ivachtchenko A. Dmitriev D. E. Golovina E. S. Kadieva M. G. Koryakova A. G. Kysil V. M. Mitkin O. D. Okun I. M. Tkachenko S. E. Vorobiev A. A. J. Med. Chem. 2010;53:5186–5196. doi: 10.1021/jm100350r. [DOI] [PubMed] [Google Scholar]
- Hao M. Li Y. Li H. Zhang S. Int. J. Mol. Sci. 2011;12:5011–5030. doi: 10.3390/ijms12085011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Vera J. A. Medina R. A. Martín-Fontecha M. Gonzalez A. De La Fuente T. Vázquez-Villa H. García-Cárceles J. Botta J. McCormick P. J. Benhamú B. Pardo L. López-Rodríguez M. L. Sci. Rep. 2017;7:41293. doi: 10.1038/srep41293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera G. Lagos C. F. Almendras S. Hebel D. Flores F. Valle-Corvalán G. Pessoa-Mahana C. D. Mella-Raipán J. Montecinos R. Recabarren-Gajardo G. Molecules. 2016;21:1070. doi: 10.3390/molecules21081070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissantz C. Kuhn B. Stahl M. J. Med. Chem. 2010;53:5061–5084. doi: 10.1021/jm100112j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell M. G. N. Baker R. J. Barden L. Beer M. S. Bristow L. Broughton H. B. Knowles M. Mcallister G. Patel S. Castro L. J. Med. Chem. 2001;44:3881–3895. doi: 10.1021/jm010943m. [DOI] [PubMed] [Google Scholar]
- Nyandege A. Kolanos R. Roth B. L. Glennon R. A. Bioorg. Med. Chem. Lett. 2007;17:1691–1694. doi: 10.1016/j.bmcl.2006.12.089. [DOI] [PubMed] [Google Scholar]
- Staroń J. Warszycki D. Kalinowska-Tłuscik J. Satała G. Bojarski A. J. RSC Adv. 2015;5:25806–25815. doi: 10.1039/C5RA00054H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groom C. R. Bruno I. J. Lightfoot M. P. Ward S. C. Acta Crystallogr. 2016;B72:171–179. doi: 10.1107/S2052520616003954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brameld K. A. Kuhn B. Reuter D. C. Stahl M. J. Chem. Inf. Model. 2008;48:1–24. doi: 10.1021/ci7002494. [DOI] [PubMed] [Google Scholar]
- Senger S. Convery M. A. Chan C. Watson N. S. Bioorg. Med. Chem. Lett. 2006;16:5731–5735. doi: 10.1016/j.bmcl.2006.08.092. [DOI] [PubMed] [Google Scholar]
- Senger S. Chan C. Convery M. A. Hubbard J. A. Shah G. P. Watson N. S. Young R. J. Bioorg. Med. Chem. Lett. 2007;17:2931–2934. doi: 10.1016/j.bmcl.2007.02.034. [DOI] [PubMed] [Google Scholar]
- Tsai Y. Dukat M. Slassi A. MacLean N. Demchyshyn L. Savage J. E. Roth B. L. Hufesein S. Lee M. Glennon R. A. Bioorg. Med. Chem. Lett. 2000;10:2295–2299. doi: 10.1016/S0960-894X(00)00453-4. [DOI] [PubMed] [Google Scholar]
- Nirogi R. V. S. Deshpande A. D. Kambhampati R. Badange R. K. Kota L. Daulatabad A. V. Shinde A. K. Ahmad I. Kandikere V. Jayarajan P. Dubey P. K. Bioorg. Med. Chem. Lett. 2011;21:346–349. doi: 10.1016/j.bmcl.2010.11.001. [DOI] [PubMed] [Google Scholar]
- Ahmed M. Briggs M. A. Bromidge S. M. Buck T. Campbell L. Deeks N. J. Garner A. Gordon L. Hamprecht D. W. Holland V. Johnson C. N. Medhurst A. D. Mitchell D. J. Moss S. F. Powles J. Seal J. T. Stean T. O. Stemp G. Thompson M. Trail B. Upton N. Winborn K. Witty D. R. Bioorg. Med. Chem. Lett. 2005;15:4867–4871. doi: 10.1016/j.bmcl.2005.06.107. [DOI] [PubMed] [Google Scholar]
- CrysAlisPro Oxford Diffraction Ltd., V. 1. 171. 36. 2., Rigaku-Oxford Diffraction, Abingdon, England, 2006 [Google Scholar]
- Sheldrick G. M. Acta Crystallogr. 2008;A64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- Farrugia L. J. J. Appl. Crystallogr. 2012;45:849–854. doi: 10.1107/S0021889812029111. [DOI] [Google Scholar]
- Macrae C. F. Bruno I. J. Chisholm J. A. Edgington P. R. McCabe P. Pidcock E. Rodriguez-Monge L. Taylor R. Van De Streek J. Wood P. A. J. Appl. Crystallogr. 2008;41:466–470. doi: 10.1107/S0021889807067908. [DOI] [Google Scholar]
- Nardelli M. J. Appl. Crystallogr. 1995;28:659–660. doi: 10.1107/S0021889895007138. [DOI] [Google Scholar]
- Keith T. A., AIMAll (Version 13.11.04), TK Gristmill Software, Overland Park KS, USA, 2013 [Google Scholar]; Espinosa E. Molins E. Lecomte C. Chem. Phys. Lett. 1998;285:170–173. doi: 10.1016/S0009-2614(98)00036-0. [DOI] [Google Scholar]
- Contreras-García J. Johnson E. R. Keinan S. Chaudret R. Piquemal J. P. Beratan D. N. Yang W. J. Chem. Theory Comput. 2011;7:625–632. doi: 10.1021/ct100641a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch M. J., Trucks G. W., Schlegel H. B., Scuseria G. E., Robb M. A., Cheeseman J. R., Scalmani G., Barone V., Mennucci B., Petersson G. A., Nakatsuji H., Caricato M., Li X., Hratchian H. P., Izmaylov A. F., Bloino J., Zheng G., Sonnenberg J. L., Hada M., Ehara M., Toyota K., Fukuda R., Hasegawa J., Ishida M., Nakajima T., Honda Y., Kitao O., Nakai H., Vreven T., Montgomery Jr. J. A., Peralta J. E., Ogliaro F., Bearpark M., Heyd J. J., Brothers E., Kudin K. N., Staroverov V. N., Kobayashi R., Normand J., Raghavachari K., Rendell A., Burant J. C., Iyengar S. S., Tomasi J., Cossi M., Rega N., Millam J. M., Klene M., Knox J. E., Cross J. B., Bakken V., Adamo C., Jaramillo J., Gomperts R., Stratmann R. E., Yazyev O., Austin A. J., Cammi R., Pomelli C., Ochterski J. W., Martin R. L., Morokuma K., Zakrzewski V. G., Voth G. A., Salvador P., Dannenberg J. J., Dapprich S., Daniels A. D., Farkas Ö., Foresman J. B., Ortiz J. V., Cioslowski J., and Fox D. J., Gaussian 09, Gaussian, Inc., Wallingford CT, 2009 [Google Scholar]
- Bader R. F. W., Atoms in Molecles. A Quantum Theory, Clarendon Press, Oxford, UK, 1990 [Google Scholar]
- Bruno I. J. Cole J. C. Edgington P. R. Kessler M. Macrae C. F. McCabe P. Pearson J. Taylor R. Acta Crystallogr. 2002;B58:389–397. doi: 10.1107/s0108768102003324. [DOI] [PubMed] [Google Scholar]
- Berman H. M. Westbrook J. Feng Z. Gilliland G. Bhat T. N. Weissig H. Shindyalov I. N. Bourne P. E. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardelli M. Comput. Chem. 1983;7:95–98. doi: 10.1016/0097-8485(83)85001-3. [DOI] [Google Scholar]
- Beddoes R. L. Dalton L. Joule J. A. Mills O. S. Street J. Watt C. I. F. J. Chem. Soc., Perkin Trans. 2. 1986:787–797. doi: 10.1039/P29860000787. [DOI] [Google Scholar]
- Chataigner I. Panel C. Gérard H. Piettre S. R. Chem. Commun. 2007;31:3288–3290. doi: 10.1039/B705034H. [DOI] [PubMed] [Google Scholar]
- Espinosa E. Molins E. Lecomte C. Chem. Phys. Lett. 1998;285:170–173. doi: 10.1016/S0009-2614(98)00036-0. [DOI] [Google Scholar]
- Lane J. R. Contreras-García J. Piquemal J. P. Miller B. J. Kjaergaard H. G. J. Chem. Theory Comput. 2013;9:3263–3266. doi: 10.1021/ct400420r. [DOI] [PubMed] [Google Scholar]
- Johnson E. R. Keinan S. Mori-Sánchez P. Contreras-García J. Cohen A. J. Yang W. JACS. 2010;132:6498–6506. doi: 10.1021/ja100936w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras-García J. Yang W. Johnson E. R. J. Phys. Chem. A. 2011;115:12983–12990. doi: 10.1021/jp204278k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano H. Yamaguchi Y. Niimi T. Sakashita T. Acta Crystallogr. 2015;D71:918–927. doi: 10.1107/S1399004715002175. [DOI] [PubMed] [Google Scholar]
- Brändén G. Sjögren T. Schnecke V. Xue Y. Drug Discov. Today. 2014;19:905–911. doi: 10.1016/j.drudis.2014.03.012. [DOI] [PubMed] [Google Scholar]
- Lu I. L. Mahindroo N. Liang P. H. Peng Y. H. Kuo C. J. Tsai K. C. Hsieh H. P. Chao Y. S. Wu S. Y. J. Med. Chem. 2006;49:5154–5161. doi: 10.1021/jm060207o. [DOI] [PubMed] [Google Scholar]
- Yan X. Wang Z. Sudom A. Cardozo M. Degraffenreid M. Di Y. Fan P. He X. Jaen J. C. Labelle M. Liu J. Ma J. McMinn D. Miao S. Sun D. Tang L. Tu H. Ursu S. Walker N. Ye Q. Powers J. P. Bioorg. Med. Chem. Lett. 2010;20:7071–7075. doi: 10.1016/j.bmcl.2010.09.097. [DOI] [PubMed] [Google Scholar]
- Coleman R. G. Carchia M. Sterling T. Irwin J. J. Shoichet B. K. PLoS One. 2013;8:e75992. doi: 10.1371/journal.pone.0075992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner R. A. Banks J. L. Murphy R. B. Halgren T. A. Klicic J. J. Mainz D. T. Repasky M. P. Knoll E. H. Shelley M. Perry J. K. Shaw D. E. Francis P. Shenkin P. S. J. Med. Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- Irwin J. J. Shoichet B. K. J. Med. Chem. 2016;59:4103–4120. doi: 10.1021/acs.jmedchem.5b02008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger, Schrodinger Suite 2016-4 MacroModel, LLC, New York, NY, 2016 [Google Scholar]
- Yunta M. J. R. AJMO. 2017;5:24–57. doi: 10.12691/ajmo-5-1-3. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.