Figure 2.
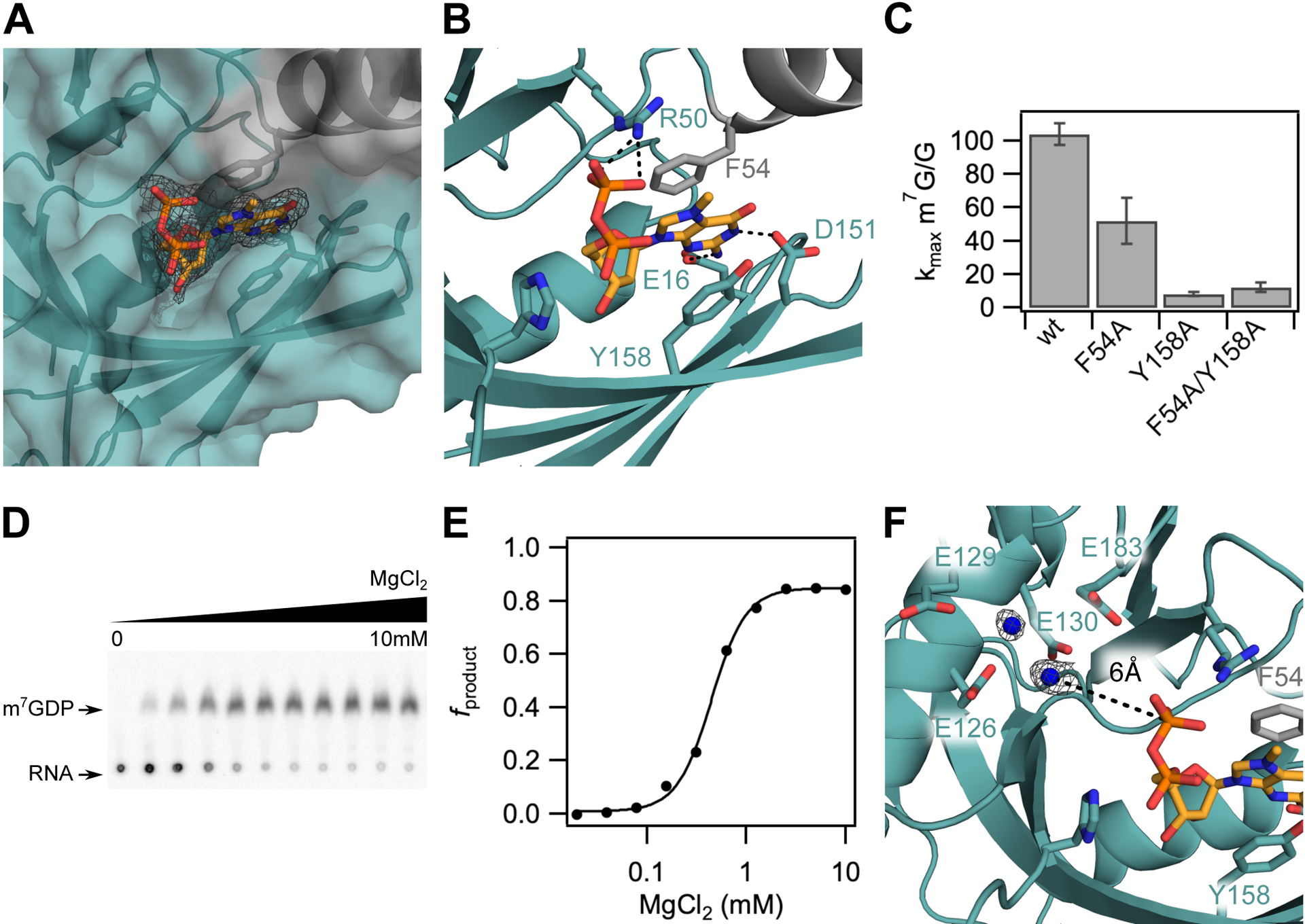
The m7G cap is positioned in a composite nucleotide binding site in D9. (A) Surface view of the m7GDP binding pocket located at the interdomain interface of the Nudix (teal) and insertion (grey) domains. Fo-Fc omit map is shown for m7GDP substrate at 2.5σ. (B) Close up view showing interactions between the m7GDP substrate and both domains of D9. The methylated guanine base is sandwiched between conserved aromatic residues F54 and Y158, and hydrogen-bonds with E16 and D151 on the sugar and Watson-Crick edges, respectively. The phosphate chain is stabilized by hydrogen-bond contacts with R50. (C) Bar graph showing methylated cap specificity (ratio of decapping rates of RNA containing methylated cap to unmethylated cap) for wild-type D9 and alanine substitutions of aromatic m7G binding residues. Error bars are s.e.m. for the rate measured in two independent experiments. (D) TLC plate monitoring cap hydrolysis of wild-type D9 at varied [MgCl2] (0 to 10mM). (E) Plot of fraction m7GDP product of wild-type D9 vs. magnesium concentration (Hill coefficient = 2.5; [Mg2+]1/2 = 0.44mM). (F) Close up view of the conserved glutamate residues in the catalytic Nudix helix. Four conserved glutamate residues (E126, E129, E130, E183) are shown as sticks. Water molecules are shown as blue spheres. Fo-Fc omit map for the waters is shown at 2.5σ in black. The distance between the β phosphate and the nearest water is 6.0 Ǻ (black dashed line).
