Figure 2. Circular RNA apparent size changes with electrophoresis conditions.
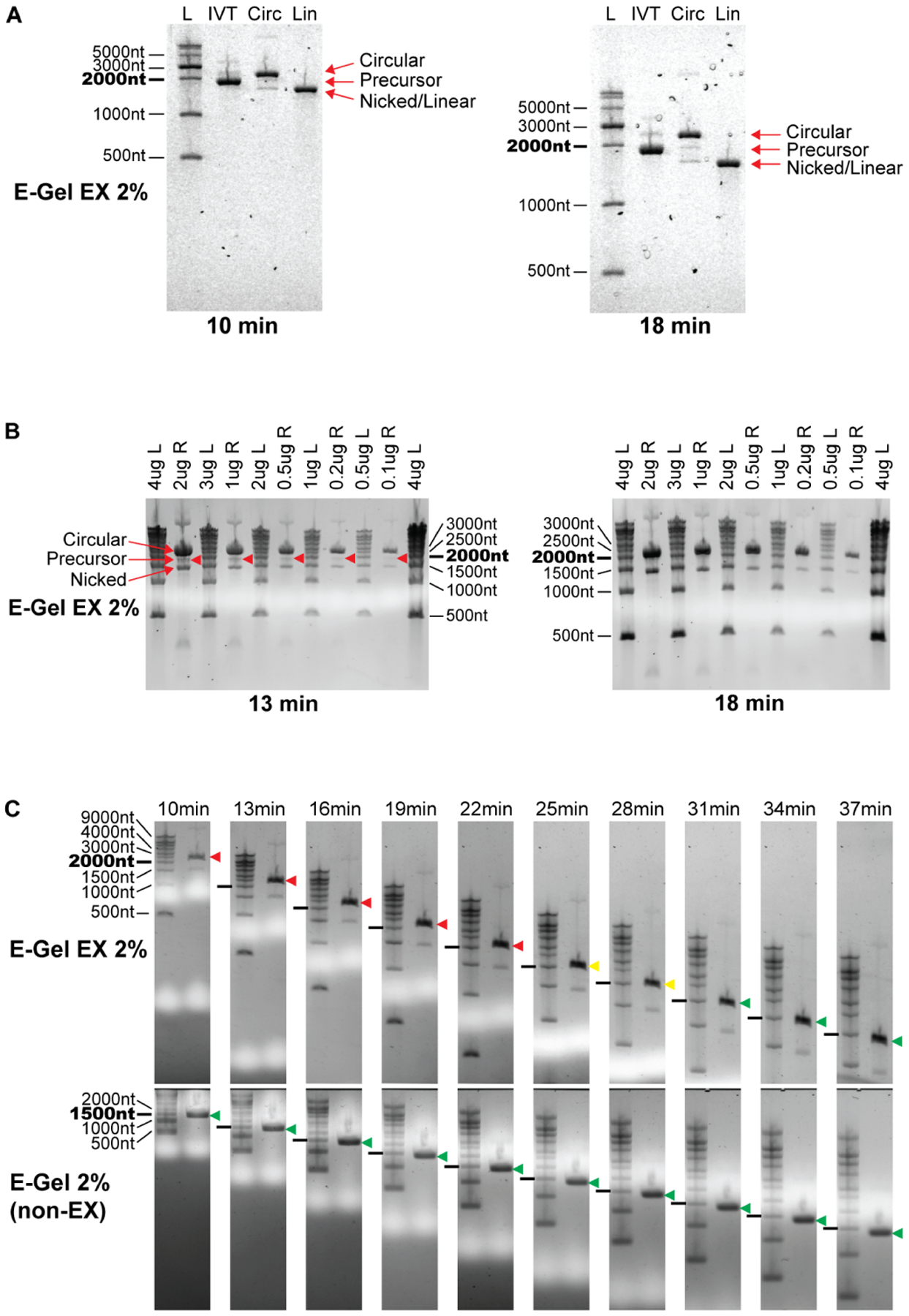
(A) Circular RNA migration on E-Gel EX systems using sample buffer containing formamide only. Circular RNA migrates slower than precursor RNA when run on E-Gel EX 2% gels using program “EX1–2%” for 10 minutes at room temperature. Extending electrophoresis duration to 18 minutes enhances separation of circular and linear splicing reaction species and maintains the same migration pattern as 10 minutes. Gel artifacts are visible as bubbles in the gel, increasing in severity with longer electrophoresis. RNA species are labeled on the right.
(B) Circular RNA migration can overlap precursor RNA when using commercially available sample buffer. Circular RNA was denatured in GLB II sample buffer and electrophoresis performed as in (A). The circular RNA band is clearly separated from the precursor band after 13 minutes of electrophoresis, however overlaps the precursor after 18 minutes. The mass of RNA loaded into the gel does not affect this migration pattern. L, Ladder; R, Circularized RNA not treated with RNaseR.
(C) Changes in circular RNA apparent size is specific to the E-Gel EX system. Top panels: E-Gel EX; bottom panels: non-EX E-Gel. RNaseR treated circularized RNA samples were denatured in GLB II buffer and subjected to the same electrophoresis conditions as in (A). Time points are indicated above gels. Circular RNA migration relative to the 2000nt RNA marker is shown by colored arrowheads (red: above 2000nt marker, yellow: with 2000nt marker, green: below 2000nt marker).
