Figure 3. Sample buffer conditions affect migration pattern of circular RNA.
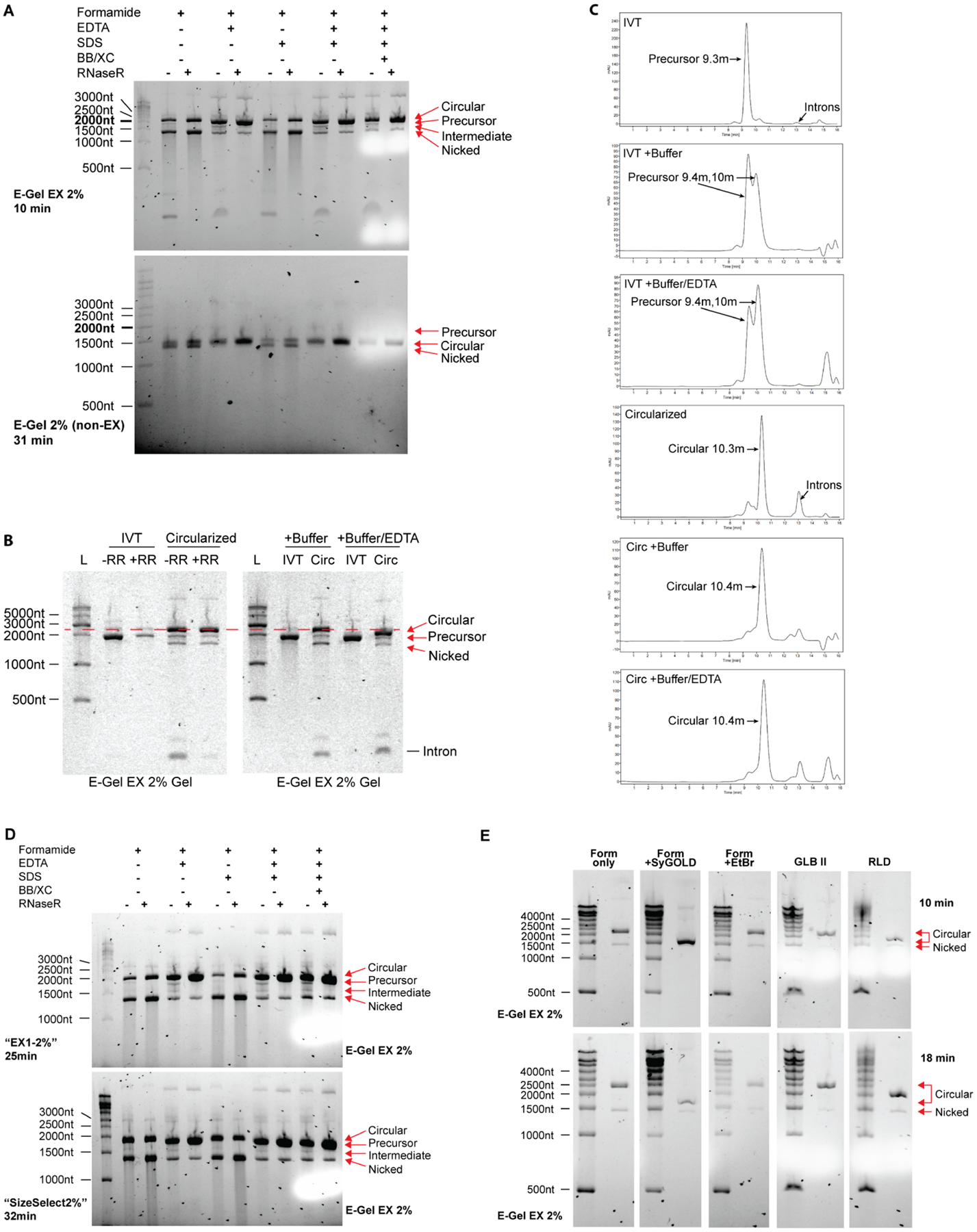
(A) EDTA decreases apparent size of circular RNA and protects circular RNA from nicking and degradation. Circularized RNA (treated with and without RNaseR) were denatured in sample buffer containing reagents depicted in the figure and run on either E-Gel EX 2% or non-EX E-Gel 2% precast gels for the indicated time using program “EX1–2%” at room temperature. Concentrations of each reagent prior to 1:1 mixing with RNA is as follows: Formamide: 95%, EDTA: 18mM, SDS: 0.025%, Bromophenol Blue (BB) and Xylene Cyanol (XC) 0.025% each. Ladder is shown on the left and was prepared in formamide only sample buffer. RNA species are labeled on the right.
(B) Buffer carryover in circular RNA preparations alters circular RNA mobility during electrophoresis in the presence of 50mM EDTA. The distance between circular RNA and linear precursor RNA narrows when 1x T4 RNA Ligase I buffer and EDTA are added to samples prior to gel loading, but not when buffer alone is added (red dotted line). A proportionally smaller shift towards lower molecular weight is visible for precursor RNA. RNA species are labeled on the right.
(C) Buffer carryover in circular RNA preparations dramatically alters precursor RNA mobility during size exclusion chromatography through the emergence of a new peak, but minimally affects circular RNA mobility. 1x T4 RNA ligase buffer I was added to samples prior to HPLC sample injection with or without 50mM EDTA. Note the lack of introns in all IVT conditions indicating that splicing is not occurring.
(D) Lower voltage causes overlap of circular RNA with precursor linear RNA. RNA was prepared in sample buffers as in (A), loaded onto two E-Gel EX 2% gels and subjected to either a high voltage program “EX1–2%” (upper) or low voltage program “SizeSelect2%” (lower) at room temperature for the time periods indicated on the left. These times are comparable with each voltage setting based on the distance of xylene cyanol migration depicted as the white areas on the right side of each gel. Ladder is shown on the left and was prepared in formamide only sample buffer. RNA species are labeled on the right.
(E) Denaturation in the presence of SYBR-GOLD I in the sample buffer abrogates the aberrant migration pattern of circular RNA seen on E-Gel EX systems. Ladder and RNaseR-treated circular RNA were denatured in the sample buffers described on top at a 1:1 ratio of sample to buffer, loaded onto E-Gel EX 2% gels, and run for 10 minutes (top panel) or 18 minutes (bottom panel) at room temperature using program “EX1–2%”. RNA species are labeled on the right. Sample buffer conditions prior to 1:1 dilution: Form only: 95% formamide; Form +SyGOLD: 95% formamide with 0.025% SYBR-GOLD I; Form +EtBr: 95% formamide with 0.025% ethidium bromide; GLB II: 95% formamide with 18mM EDTA and 0.025% each of SDS, bromophenol blue, and xylene cyanol; RLD: 95% formamide with 0.5mM EDTA and 0.025% each of SDS, ethidium bromide, bromophenol blue, and xylene cyanol.
